Search Count: 31
 |
Crystal Structures Of Branched-Chain Aminotransferase From Deinococcus Radiodurans Complexes With Alpha-Ketoisocaproate And L-Glutamate Suggest Its Radio-Resistance For Catalysis
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-12-12 Classification: TRANSFERASE Ligands: PLP |
 |
Crystal Structures Of Branched-Chain Aminotransferase From Deinococcus Radiodurans Complexes With Alpha-Ketoisocaproate And L-Glutamate Suggest Its Radio-Resistance For Catalysis
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2012-12-12 Classification: TRANSFERASE Ligands: PLP, COI |
 |
Crystal Structures Of Branched-Chain Aminotransferase From Deinococcus Radiodurans Complexes With Alpha-Ketoisocaproate And L-Glutamate Suggest Its Radio-Resistance For Catalysis
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-12-12 Classification: TRANSFERASE Ligands: PLP, GLU |
 |
Organism: Desulfovibrio gigas
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2010-09-22 Classification: OXIDOREDUCTASE Ligands: SO3, SRM, SF4 |
 |
Organism: Desulfovibrio gigas
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2010-09-22 Classification: OXIDOREDUCTASE Ligands: SF4, SO3, SRM |
 |
Crystal Structure Of The Complex Of Pdz-Rhogef Dh/Ph Domains With Gtp-Gamma-S Activated Rhoa
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2010-04-28 Classification: SIGNALING PROTEIN Ligands: MG, GSP |
 |
High Resolution Crystal Structure Of Bacillus Amyloliquefaciens Alpha-Amylase
Organism: Bacillus amyloliquefaciens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2008-12-09 Classification: HYDROLASE Ligands: CA, NA |
 |
Identification And Characterization Of Two Amino Acids Critical For The Substrate Inhibition Of Sult2A1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2008-07-29 Classification: TRANSFERASE Ligands: AOX |
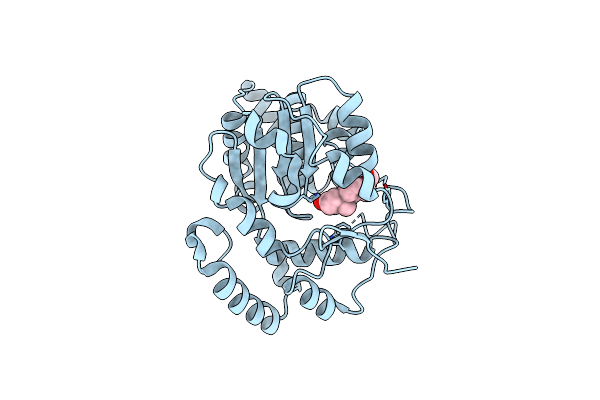 |
Identification And Characterization Of Two Amino Acids Critical For The Substrate Inhibition Of Sult2A1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2008-07-29 Classification: TRANSFERASE Ligands: AOX |
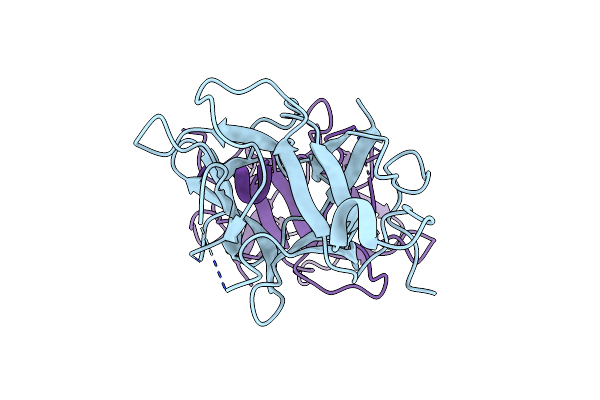 |
Structure And Function Study Of Rice Bifunctional Alpha-Amylase/Subtilisin Inhibitor From Oryza Sativa
Organism: Oryza sativa subsp. japonica
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2008-07-22 Classification: HYDROLASE INHIBITOR |
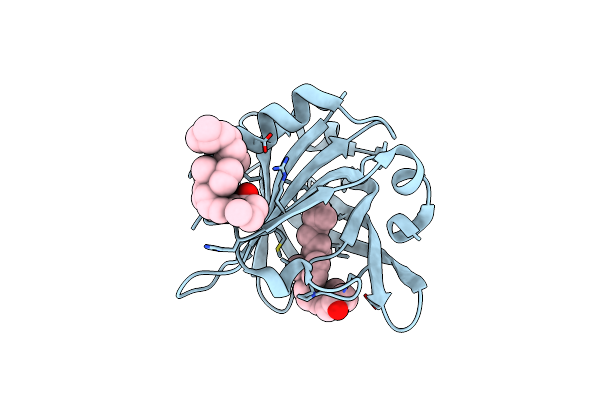 |
Crystal Structure Of A Secondary Vitamin D3 Binding Site Of Milk Beta-Lactoglobulin
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2007-10-02 Classification: TRANSPORT PROTEIN Ligands: VD3 |
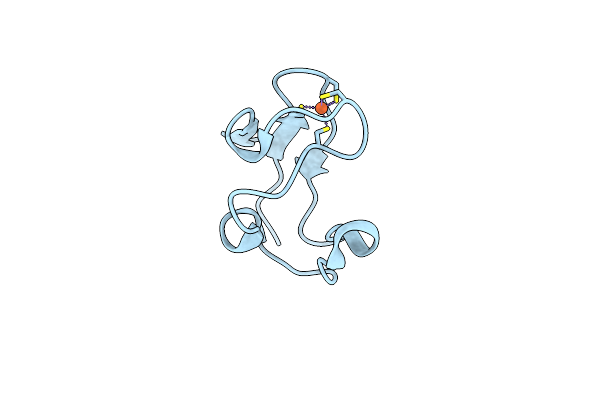 |
Crystal Structure Of Rubredoxin From Desulfovibrio Gigas To Ultra-High 0.68 A Resolution
Organism: Desulfovibrio gigas
Method: X-RAY DIFFRACTION Resolution:0.68 Å Release Date: 2006-10-10 Classification: ELECTRON TRANSPORT Ligands: FE |
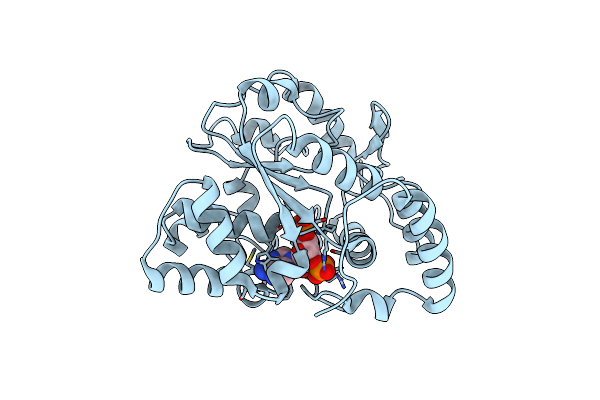 |
Crystal Structures Of Sult1A2 And Sult1A1*3: Implications In The Bioactivation Of N-Hydroxy-2-Acetylamino Fluorine (Oh-Aaf)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2006-05-30 Classification: TRANSFERASE Ligands: A3P |
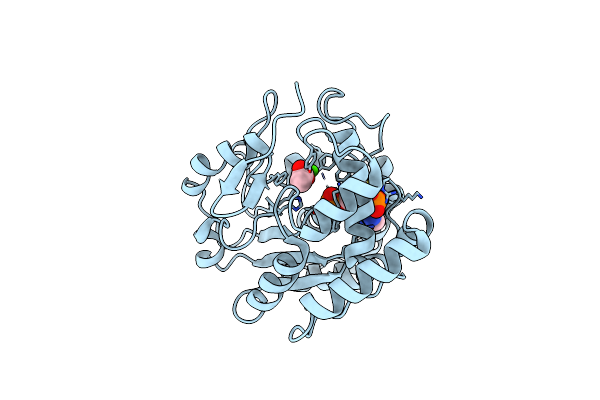 |
Crystal Structures Of Sult1A2 And Sult1A1*3: Implications In The Bioactivation Of N-Hydroxy-2-Acetylamino Fluorine (Oh-Aaf)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2006-05-30 Classification: TRANSFERASE Ligands: CA, A3P, ACY |
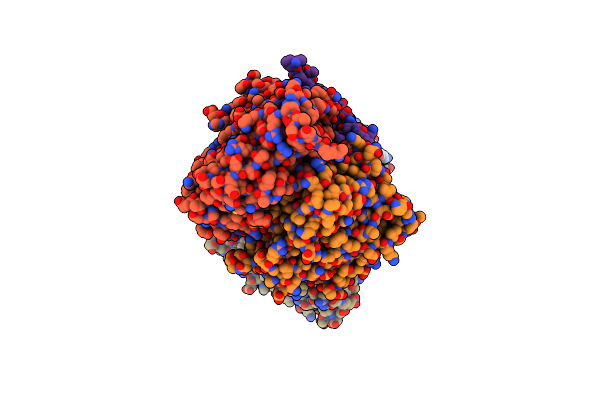 |
Crystal Structure Of C-Terminal Desundecapeptide Nitrite Reductase From Achromobacter Cycloclastes
Organism: Achromobacter cycloclastes
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2005-12-20 Classification: OXIDOREDUCTASE Ligands: CU, CL |
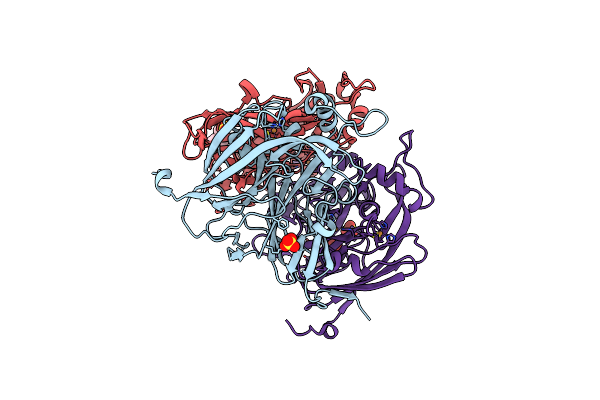 |
Crystal Structure Of C-Terminal Despentapeptide Nitrite Reductase From Achromobacter Cycloclastes At Ph6.2
Organism: Achromobacter cycloclastes
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2004-03-30 Classification: OXIDOREDUCTASE Ligands: CU, SO4, MES |
 |
Crystal Structure Of C-Terminal Despentapeptide Nitrite Reductase From Achromobacter Cycloclastes At Ph5.0
Organism: Achromobacter cycloclastes
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2004-03-30 Classification: OXIDOREDUCTASE Ligands: CU, SO4, ACY |
 |
Crystal Structure Of A No-Forming Nitrite Reductase Mutant: An Analog Of A Transition State In Enzymatic Reaction
Organism: Achromobacter cycloclastes
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2003-11-04 Classification: OXIDOREDUCTASE Ligands: CU |
 |
Reduced Hybrid Cluster Protein From Desulfovibrio Desulfuricans X-Ray Structure At 1.25A Resolution
Organism: Desulfovibrio desulfuricans
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2003-04-08 Classification: HYBRID CLUSTER PROTEIN Ligands: SF4, SF3, MES |
 |
Reduced Hybrid Cluster Protein (Hcp) From Desulfovibrio Vulgaris Hildenborough Structure At 1.55A Resolution Using Synchrotron Radiation.
Organism: Desulfovibrio vulgaris
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2003-04-08 Classification: OXIDOREDUCTASE Ligands: SF4, SF3, GOL |

