Search Count: 18
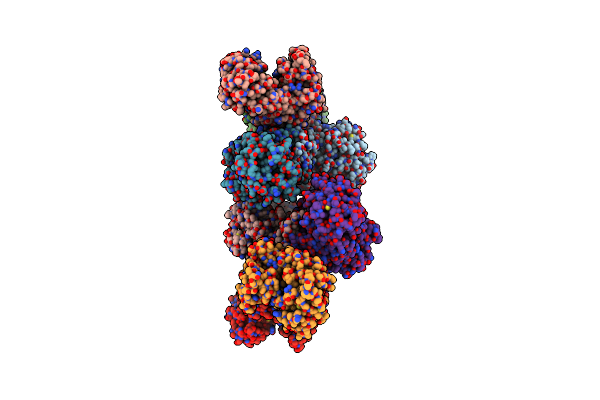 |
Complex Structure Of A Glucose 6-Phosphate Dehydrogenase From Zymomonas Mobilis
Organism: Zymomonas mobilis
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2023-04-12 Classification: OXIDOREDUCTASE Ligands: NMN |
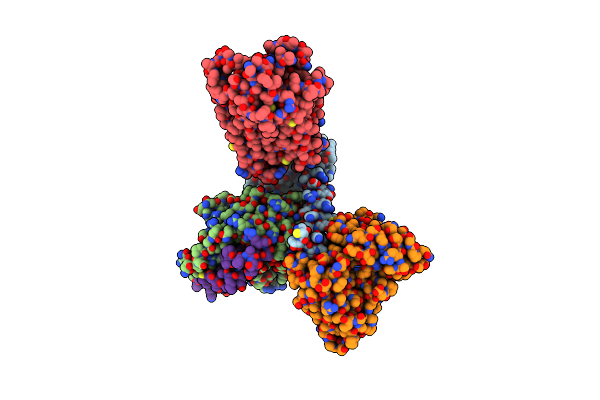 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-15 Classification: MEMBRANE PROTEIN Ligands: 7NR |
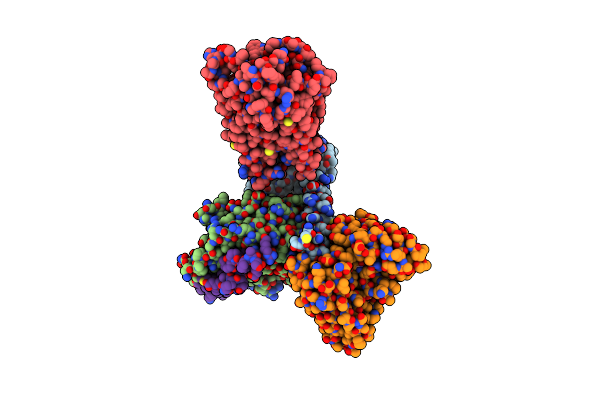 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-15 Classification: MEMBRANE PROTEIN Ligands: EIC |
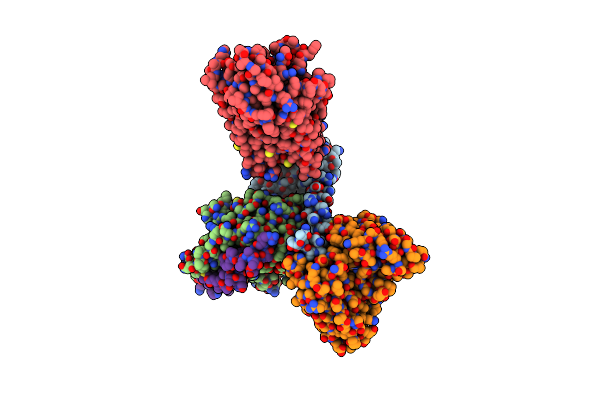 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-15 Classification: MEMBRANE PROTEIN Ligands: OLA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-15 Classification: MEMBRANE PROTEIN Ligands: YN9 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-15 Classification: MEMBRANE PROTEIN Ligands: EPA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-08 Classification: MEMBRANE PROTEIN Ligands: YN9 |
 |
Organism: Zymomonas mobilis
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2023-02-22 Classification: OXIDOREDUCTASE |
 |
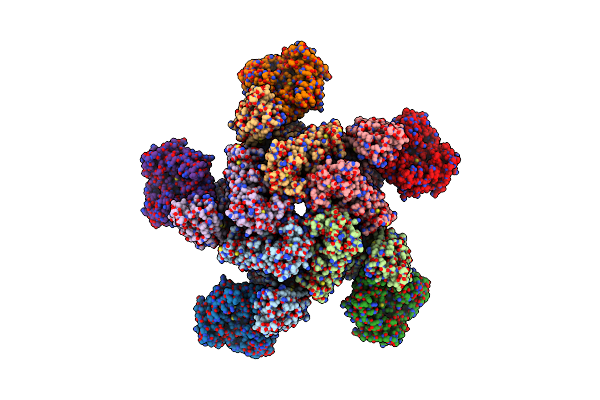
Organism: Triticum monococcum, Puccinia graminis f. sp. tritici
Method: ELECTRON MICROSCOPY Release Date: 2022-11-02 Classification: PLANT PROTEIN Ligands: ATP |
 |
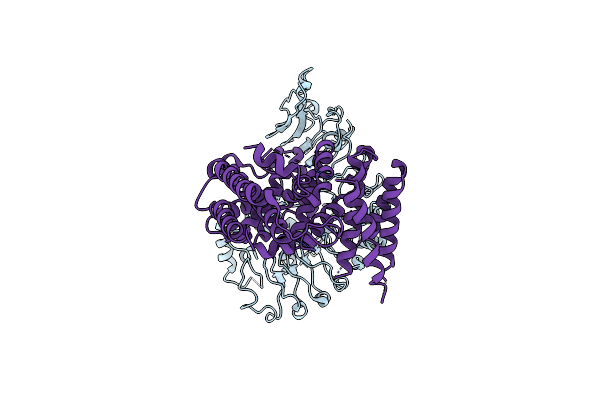
Organism: Puccinia graminis f. sp. tritici
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2022-09-28 Classification: PROTEIN TRANSPORT |
 |
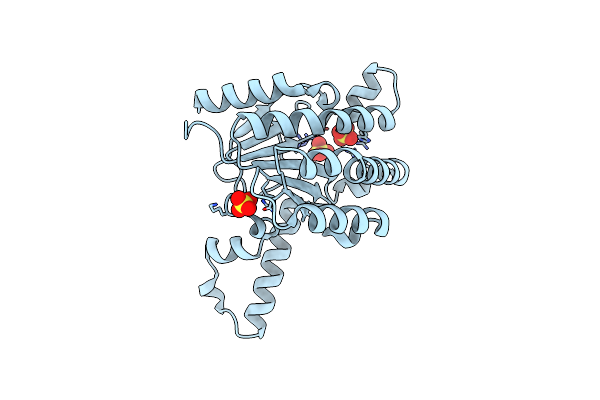
Organism: Triticum monococcum, Puccinia graminis f. sp. tritici
Method: ELECTRON MICROSCOPY Release Date: 2022-09-28 Classification: PLANT PROTEIN Ligands: ATP |
 |
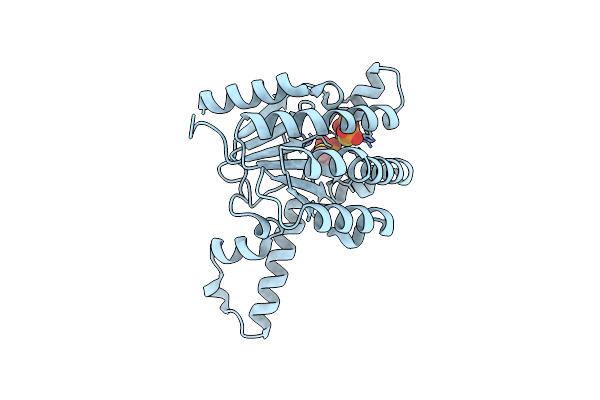
Organism: Triticum monococcum, Puccinia graminis f. sp. tritici
Method: ELECTRON MICROSCOPY Release Date: 2022-09-28 Classification: PLANT PROTEIN |
 |
Crystal Structure Of Lavandulyl Diphosphate Synthase From Lavandula X Intermedia In Apo Form
Organism: Lavandula lanata
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2016-03-02 Classification: TRANSFERASE Ligands: SO4 |
 |
Crystal Structure Of Lavandulyl Diphosphate Synthase From Lavandula X Intermedia In Complex With S-Thiolo-Isopentenyldiphosphate
Organism: Lavandula lanata
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2016-03-02 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: MG, DST |
 |
Crystal Structure Of Lavandulyl Diphosphate Synthase From Lavandula X Intermedia In Complex With Dimethylallyl Diphosphate
Organism: Lavandula lanata
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2016-03-02 Classification: TRANSFERASE Ligands: MG, PIS, 61G, SO4, ISY |
 |
Crystal Structure Of Iridoid Synthase From Cantharanthus Roseus In Complex With Nadph
Organism: Catharanthus roseus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-11-04 Classification: OXIDOREDUCTASE Ligands: NDP |
 |
Crystal Structure Of Iridoid Synthase From Cantharanthus Roseus In Complex With Nad+
Organism: Catharanthus roseus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2015-11-04 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Crystal Structure Of Iridoid Synthase From Cantharanthus Roseus In Complex With Nad+ And 10-Oxogeranial
Organism: Catharanthus roseus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2015-11-04 Classification: OXIDOREDUCTASE Ligands: NAD, XOG |

