Search Count: 51
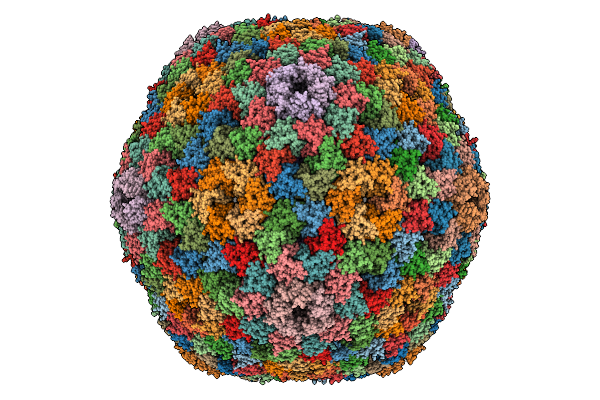 |
Organism: Halothiobacillus neapolitanus c2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: STRUCTURAL PROTEIN |
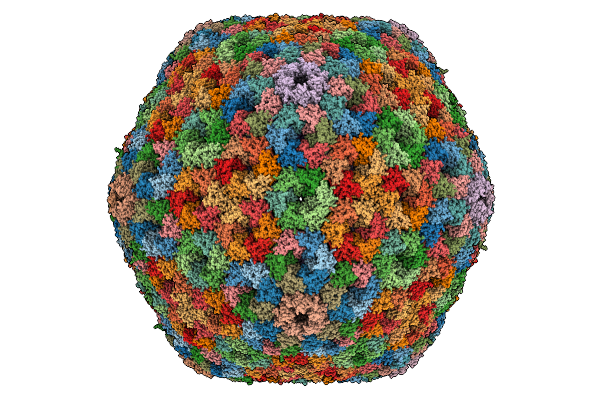 |
Cryo-Em Structure Of Carboxysomal Mid-Shell: T = 16 Shell Under C1 Symmetry.
Organism: Halothiobacillus neapolitanus c2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: STRUCTURAL PROTEIN |
 |
The Psi3-Isia43 Complex With A Closed Double Ring Of Isia Proteins Bound To A Trimeric Psi Core
Organism: Thermosynechococcus vestitus bp-1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: PHOTOSYNTHESIS Ligands: CLA, PQN, SF4, BCR, LHG, LMU, SQD, LMG, CA |
 |
The Psi1-Isia13 Complex With Double-Layered Isia Proteins Bound To The Monomeric Psi Core
Organism: Thermosynechococcus vestitus bp-1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: PHOTOSYNTHESIS Ligands: CLA, BCR, SQD, LMG, LHG, PQN, SF4, LMU |
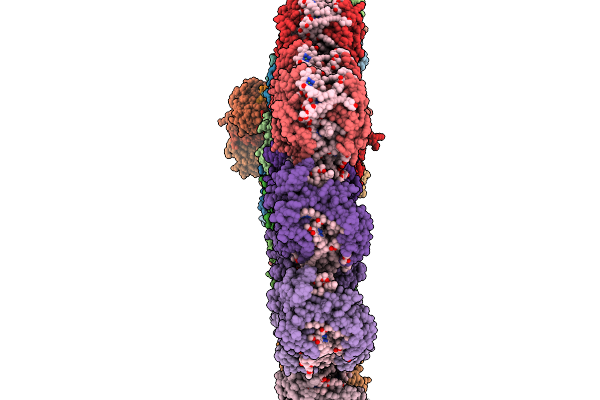 |
Organism: Emiliania huxleyi
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: PHOTOSYNTHESIS Ligands: CLA, KC2, DD6, LMG, A86, LHG, SQD, A1EB1, A1EB4, BCR, PQN, SF4, DGD |
 |
Cryo-Em Structure Of Halothiobacillus Neapolitanus Alpha-Carboxysome T=4 Mini-Shell Containing Ctd Truncated Mutant Of Csosca
Organism: Halothiobacillus neapolitanus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: STRUCTURAL PROTEIN |
 |
Cryo-Em Structure Of Alpha-Carboxysome T=4 Mini-Shell Containing Ctd Only Mutant Of Csosca
Organism: Halothiobacillus neapolitanus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: STRUCTURAL PROTEIN |
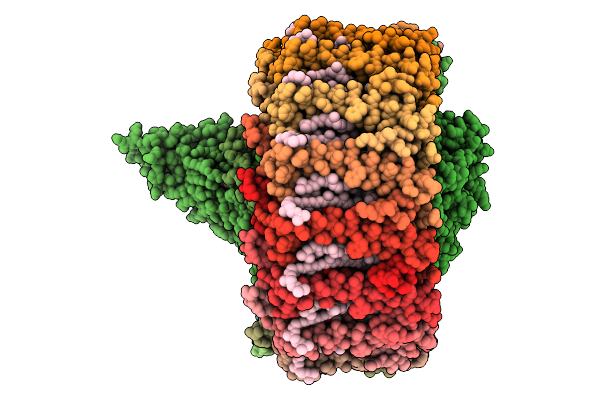 |
Organism: Dinoroseobacter shibae dfl 12 = dsm 16493
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: PHOTOSYNTHESIS Ligands: U10, BCL, SPN, MW9, HEM, BPH, FE |
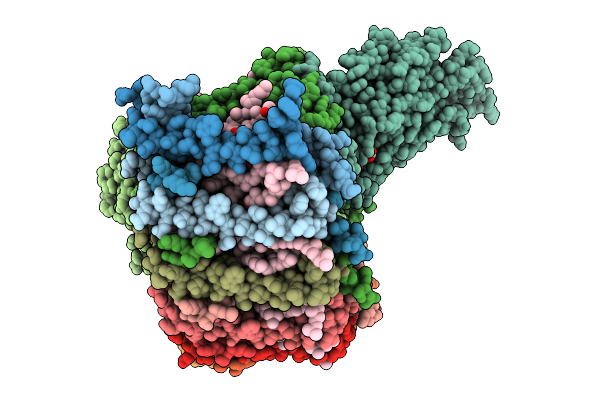 |
Cryo-Em Structure Of The Dinoroseobacter Shibae Rc-Lh1 Supercomplex With Incomplete Lh1 Ring(State 1)
Organism: Dinoroseobacter shibae dfl 12 = dsm 16493
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: PHOTOSYNTHESIS Ligands: BCL, SPN, HEM, U10, BPH, FE |
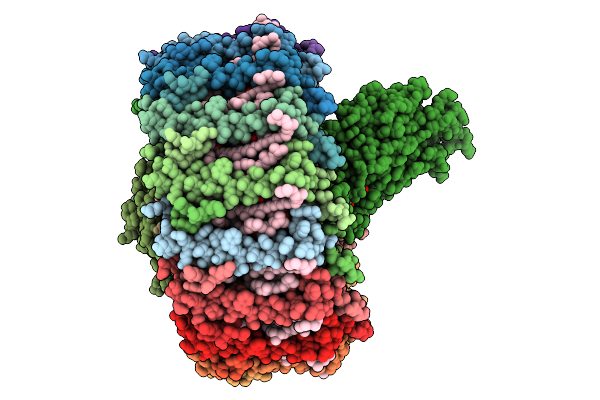 |
Cryo-Em Structure Of The Dinoroseobacter Shibae Rc-Lh1 Supercomplex With Incomplete Lh1 Ring(State 2)
Organism: Dinoroseobacter shibae dfl 12 = dsm 16493
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: PHOTOSYNTHESIS Ligands: BCL, SPN, HEM, U10, BPH, FE |
 |
Beta Carbonic Anhydrase Csosca From The Halothiobacillus Neapolitanus Alpha-Carboxysome
Organism: Streptococcus sp. 'group g', Halothiobacillus neapolitanus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: LYASE Ligands: ZN |
 |
Organism: Synechococcus elongatus pcc 7942 = fachb-805
Method: ELECTRON MICROSCOPY Release Date: 2025-05-07 Classification: PHOTOSYNTHESIS |
 |
Organism: Fuscovulum blasticum dsm 2131
Method: ELECTRON MICROSCOPY Resolution:2.84 Å Release Date: 2025-03-05 Classification: PHOTOSYNTHESIS Ligands: BCL, SPO, PC1, CDL, BPH, U10, FE2 |
 |
Organism: Fuscovulum blasticum dsm 2131
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: PHOTOSYNTHESIS Ligands: BCL, SPO, PC1, CDL, BPH, U10, FE2 |
 |
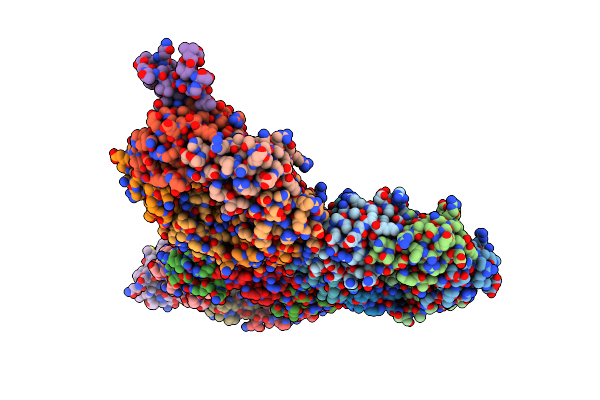
Organism: Isochrysis galbana
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: PHOTOSYNTHESIS Ligands: CLA, KC2, DD6, LHG, LMG, A86, LMU, SQD, DGD, BCR, SF4, PQN |
 |
Cryo-Em Structure Of Carboxysomal Midi-Shell:Icosahedral Assembly From Csos4A/4B/1A/1B/1C/1D And Csos2 C-Terminal Co-Expression (T = 19)
Organism: Halothiobacillus neapolitanus
Method: ELECTRON MICROSCOPY Release Date: 2024-12-11 Classification: STRUCTURAL PROTEIN |
 |
Cryo-Em Structure Of Carboxysomal Midi-Shell: Icosahedral Assembly From Csos4A/4B/1A/1B/1C/1D And Csos2 C-Terminal Co-Expression (T = 16)
Organism: Halothiobacillus neapolitanus
Method: ELECTRON MICROSCOPY Release Date: 2024-12-11 Classification: STRUCTURAL PROTEIN |
 |
Cryo-Em Structure Of Carboxysomal Midi-Shell: Icosahedral Assembly From Csos4A/4B/1A/1B/1C/1D And Csos2 C-Terminal Co-Expression (T = 9)
Organism: Halothiobacillus neapolitanus
Method: ELECTRON MICROSCOPY Release Date: 2024-12-11 Classification: STRUCTURAL PROTEIN |
 |
Cryo-Em Structure Of Carboxysomal Midi-Shell: Assembly From Csos4A/4B/1A/1B/1C/1D And Csos2 C-Terminal Co-Expression (T=9 Q=12)
Organism: Halothiobacillus neapolitanus
Method: ELECTRON MICROSCOPY Release Date: 2024-12-11 Classification: STRUCTURAL PROTEIN |
 |
Cryo-Em Structure Of Carboxysomal Midi-Shell: Icosahedral Assembly From Csos4A/4B/1A/1B/1C/1D And Csos2 C-Terminal Co-Expression (T = 13)
Organism: Halothiobacillus neapolitanus
Method: ELECTRON MICROSCOPY Release Date: 2024-12-11 Classification: STRUCTURAL PROTEIN |

