Search Count: 1,635
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: TRANSPORT PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: TRANSPORT PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: TRANSPORT PROTEIN Ligands: GLP |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: TRANSPORT PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: TRANSPORT PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: TRANSPORT PROTEIN Ligands: PO4 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: TRANSPORT PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: TRANSPORT PROTEIN Ligands: PO4 |
 |
Cryo-Em Structure Of Rnf20/Rnf40-Rad6A-Ub In Complex With H2Bs112Glcnac Nucleosome
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: NUCLEAR PROTEIN Ligands: ZN, NAG |
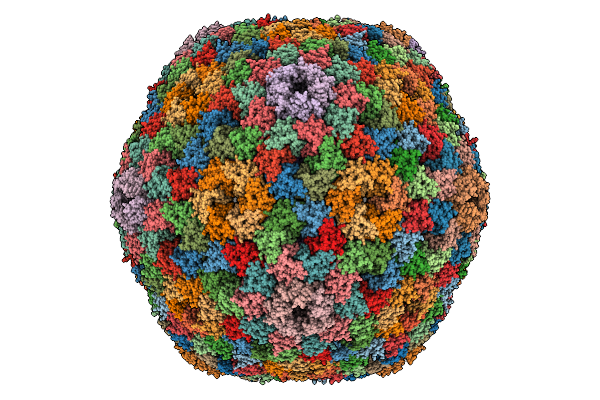 |
Organism: Halothiobacillus neapolitanus c2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: STRUCTURAL PROTEIN |
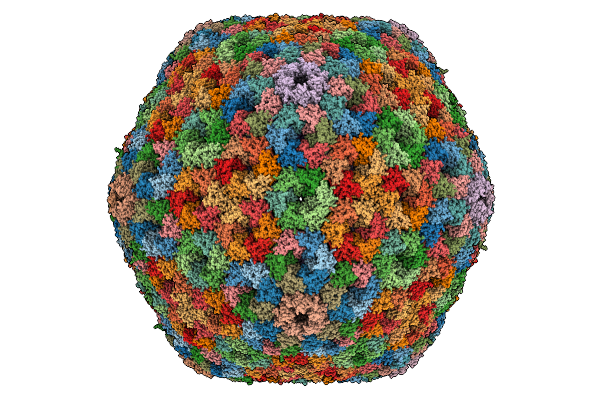 |
Cryo-Em Structure Of Carboxysomal Mid-Shell: T = 16 Shell Under C1 Symmetry.
Organism: Halothiobacillus neapolitanus c2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: STRUCTURAL PROTEIN |
 |
The Psi3-Isia43 Complex With A Closed Double Ring Of Isia Proteins Bound To A Trimeric Psi Core
Organism: Thermosynechococcus vestitus bp-1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: PHOTOSYNTHESIS Ligands: CLA, PQN, SF4, BCR, LHG, LMU, SQD, LMG, CA |
 |
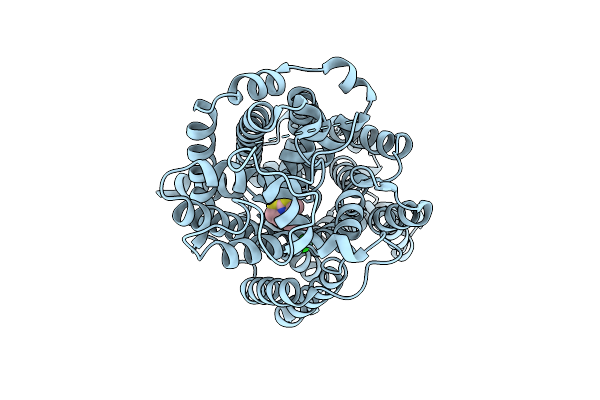
The Psi1-Isia13 Complex With Double-Layered Isia Proteins Bound To The Monomeric Psi Core
Organism: Thermosynechococcus vestitus bp-1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: PHOTOSYNTHESIS Ligands: CLA, BCR, SQD, LMG, LHG, PQN, SF4, LMU |
 |
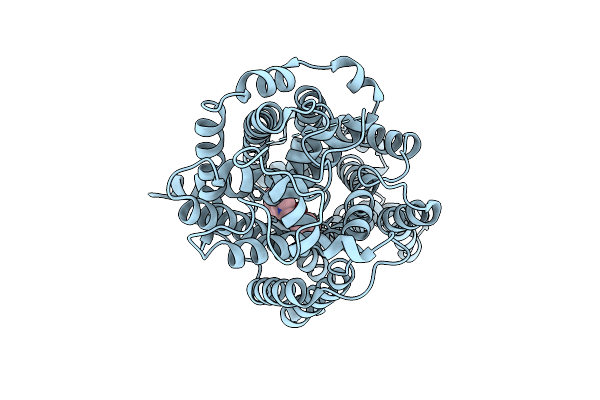
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: MEMBRANE PROTEIN Ligands: A1EBN, CL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: MEMBRANE PROTEIN Ligands: A1EBO, CL |
 |
 |
Crystal Structure Of A Glyceraldehyde-3-Phosphate Dehydrogenase From Neisseria Gonorrhoeae In Complex With Nad (P1 Form2)
Organism: Neisseria gonorrhoeae nccp11945
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: OXIDOREDUCTASE Ligands: NAD, NA |
 |
Crystal Structure Of An Iole Protein From Brucella Melitensis (Hexagonal P Form)
Organism: Brucella melitensis biotype 1 (strain atcc 23456 / ccug 17765 / nctc 10094 / 16m)
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: BIOSYNTHETIC PROTEIN Ligands: MN |
 |
Crystal Structure Of An Iole Protein From Brucella Melitensis (Orthorhombic P Form)
Organism: Brucella melitensis biotype 1 (strain atcc 23456 / ccug 17765 / nctc 10094 / 16m)
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: BIOSYNTHETIC PROTEIN Ligands: MN |
 |
Crystal Structure Of A Iole Protein From Brucella Melitensis (Orthorhombic P Form 2)
Organism: Brucella melitensis biotype 1 (strain atcc 23456 / ccug 17765 / nctc 10094 / 16m)
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: BIOSYNTHETIC PROTEIN Ligands: MN |

