Search Count: 487
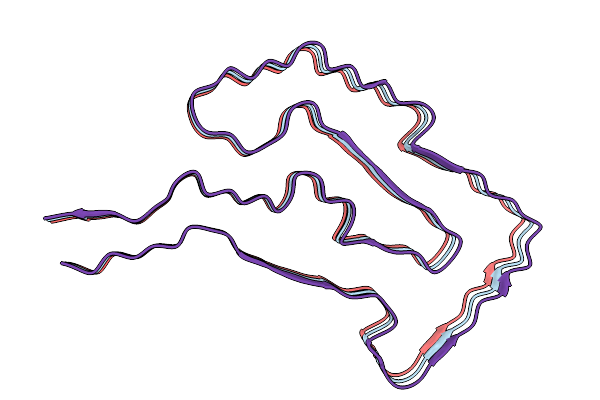 |
The Cryo-Em Structure Of Amyloid Fibrils From Abdominal Fat Of An Al Amyloidosis Patient (Case 1) - Polymorph 1.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: PROTEIN FIBRIL |
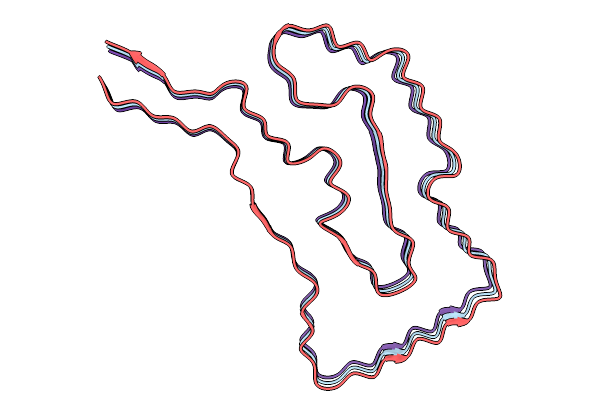 |
The Cryo-Em Structure Of Amyloid Fibrils From Heart Of An Al Amyloidosis Patient (Case 1) - Polymorph 1.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: PROTEIN FIBRIL |
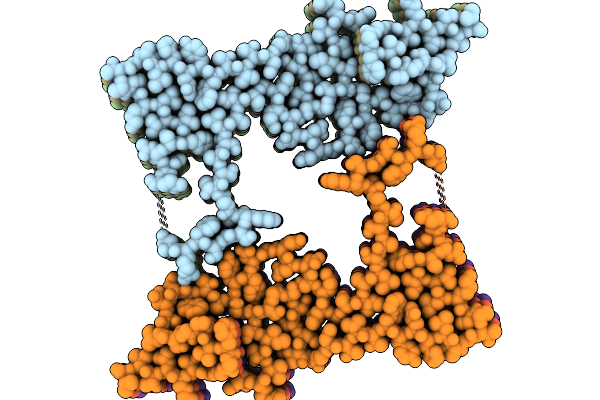 |
The Cryo-Em Structure Of Amyloid Fibrils From Abdominal Fat Of An Al Amyloidosis Patient (Case 2) - Polymorph 1.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: PROTEIN FIBRIL |
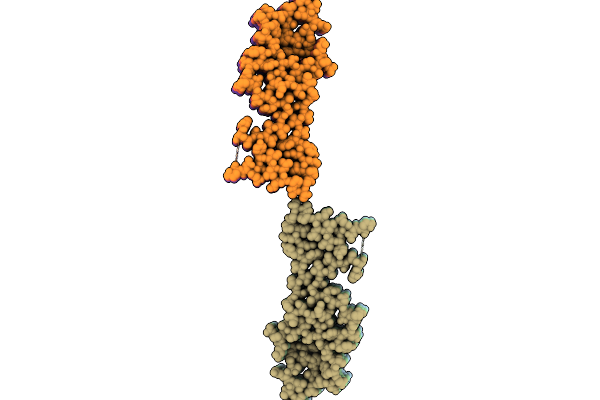 |
The Cryo-Em Structure Of Amyloid Fibrils From Abdominal Fat Of An Al Amyloidosis Patient (Case 2) - Polymorph 2.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: PROTEIN FIBRIL |
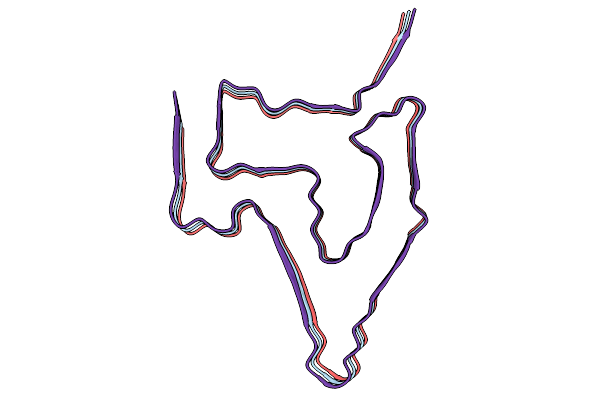 |
The Cryo-Em Structure Of Amyloid Fibrils From Abdominal Fat Of An Al Amyloidosis Patient (Case 3).
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: PROTEIN FIBRIL |
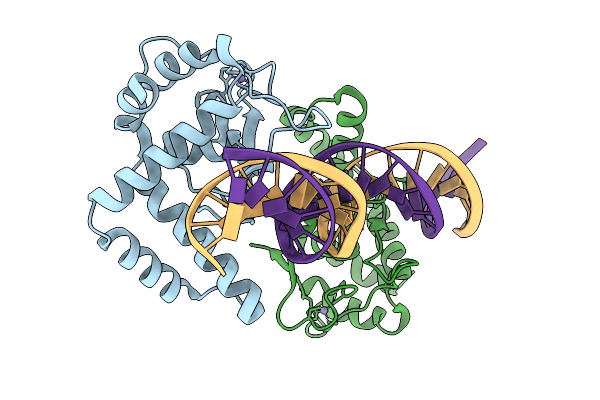 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2026-01-21 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2026-01-14 Classification: TRANSFERASE Ligands: GOL, MN, AMP, SO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2026-01-14 Classification: TRANSFERASE Ligands: AMP, SO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2026-01-14 Classification: LYASE Ligands: NAD, SO4 |
 |
Organism: Thermoproteus sp. az2
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2026-01-14 Classification: HYDROLASE Ligands: BGC, GOL, ACT |
 |
Organism: Thermoproteus sp. az2
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2026-01-14 Classification: HYDROLASE |
 |
Organism: Thermoproteus sp. az2
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2026-01-14 Classification: HYDROLASE Ligands: GOL, ACY, ACT |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: PROTEIN FIBRIL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: PROTEIN FIBRIL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: PROTEIN FIBRIL |
 |
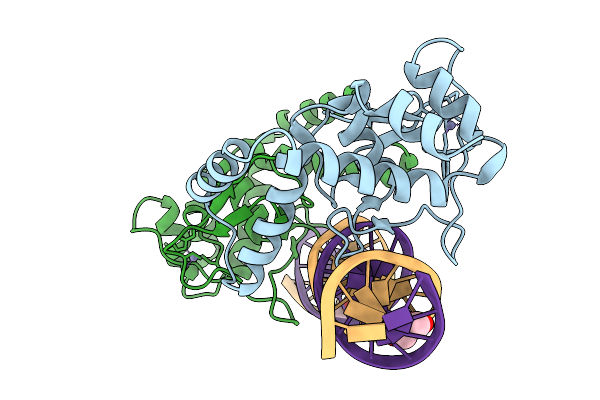
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2025-12-31 Classification: DNA BINDING PROTEIN/DNA Ligands: EDO, ZN |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2025-12-31 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN, GOL |
 |
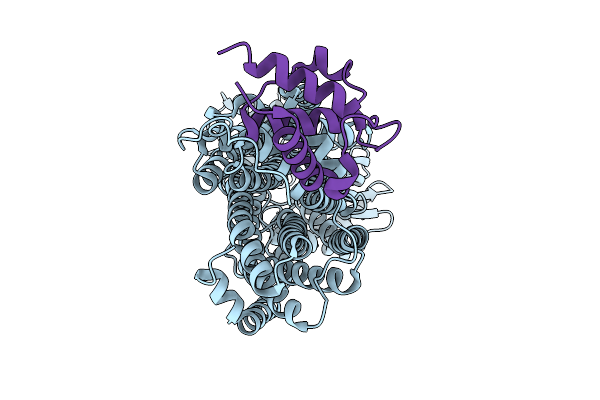
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2025-12-24 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN |
 |
Cryo-Em Map Of Msmeg_3496 In Complex With Acpm, Size Exclusion Chromatography Peak1
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN |
 |
Cryo-Em Map Of Msmeg_3496 In Complex With Acpm, Size Exclusion Chromatography Peak2
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN |

