Search Count: 38
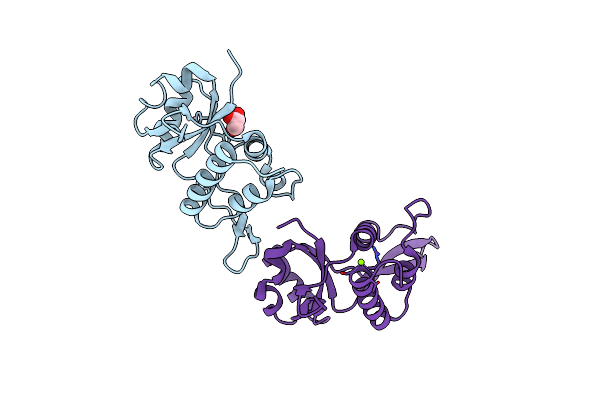 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-03-19 Classification: TRANSCRIPTION Ligands: GOL, MG |
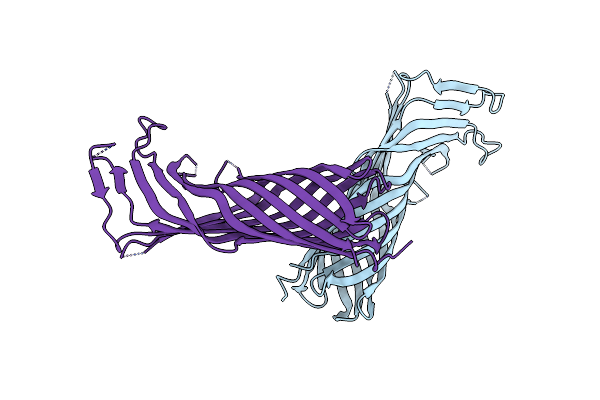 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2023-05-31 Classification: HYDROLASE Ligands: HOH |
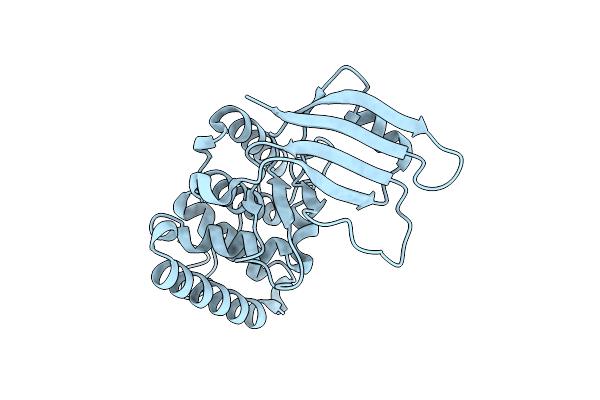 |
Crystal Structure Of The Type Iii Effector Xopai From Xanthomonas Axonopodis Pv. Citri In Space Group P41212
Organism: Xanthomonas citri
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2019-10-30 Classification: CELL INVASION |
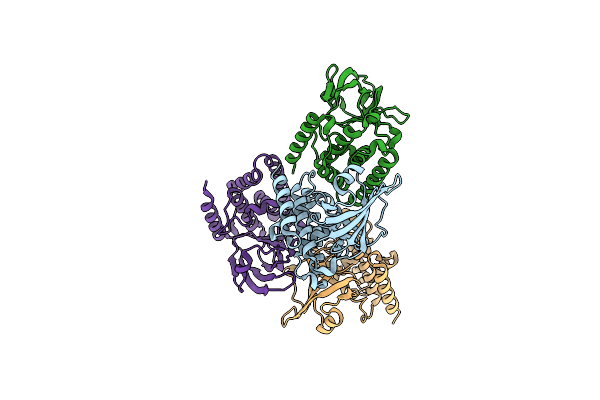 |
Crystal Structure Of The Type Iii Effector Xopai From Xanthomonas Axonopodis Pv. Citri - A 70 Residue N-Terminal Truncation
Organism: Xanthomonas citri
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2019-10-30 Classification: CELL INVASION |
 |
Crystal Structure Of The Type Iii Effector Xopai From Xanthomonas Axonopodis Pv. Citri In Space Group P43212
Organism: Xanthomonas citri
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2019-08-21 Classification: CELL INVASION |
 |
The Crystal Structure Of Human Cytosolic Nadp(+)-Dependent Malic Enzyme In Apo Form
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2014-08-13 Classification: OXIDOREDUCTASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2012-05-30 Classification: CELL ADHESION Ligands: NGA, NAG, FUC, CA |
 |
V Beta/V Beta Homodimerization-Based Pre-Tcr Model Suggested By Tcr Beta Crystal Structures
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2011-03-16 Classification: IMMUNE SYSTEM |
 |
V Beta/V Beta Homodimerization-Based Pre-Tcr Model Suggested By Tcr Beta Crystal Structures
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-03-09 Classification: IMMUNE SYSTEM Ligands: GOL, PEG, EPE |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2011-03-02 Classification: APOPTOSIS Ligands: NAG, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2009-09-22 Classification: IMMUNE SYSTEM |
 |
Crystal Structure Of Intermediate Affinity I Domain Of Integrin Lfa-1 With The Fab Fragment Of Its Antibody Al-57
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2009-09-22 Classification: cell adhesion/immune system Ligands: MN, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2009-06-23 Classification: CELL ADHESION Ligands: GOL, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2009-06-23 Classification: CELL ADHESION Ligands: ICF, GOL, MG |
 |
Xc1028 From Xanthomonas Campestris Adopts A Pilz Domain-Like Structure Yet With Trivial C-Di-Gmp Binding Activity
Organism: Xanthomonas campestris pv. campestris
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2009-05-19 Classification: UNKNOWN FUNCTION |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2008-08-26 Classification: CELL ADHESION |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2008-08-19 Classification: Cell adhesion, immune system Ligands: MG, NAG, GOL |
 |
Organism: Influenza a virus
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2008-05-27 Classification: VIRAL PROTEIN, RNA BINDING PROTEIN |
 |
Crystal Structure Of The Thrombospondin-1 N-Terminal Domain In Complex With Fractionated Heparin Dp10
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2008-01-08 Classification: CELL ADHESION Ligands: SO4 |
 |
The Crystal Structure Of The Thrombospondin-1 N-Terminal Domain In Complex With Fractionated Heparin Dp8
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2008-01-08 Classification: CELL ADHESION |

