Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Shewanella oneidensis mr-1
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: STRUCTURAL PROTEIN |
 |
Organism: Shewanella oneidensis mr-1
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: CELL ADHESION |
 |
Organism: Shewanella oneidensis mr-1
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: PROTEIN FIBRIL |
 |
The Cryo-Em Structure Of Amyloid Fibrils From Abdominal Fat Of A Multiple Myeloma Patient (Case 1)- Polymorph 1.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: PROTEIN FIBRIL |
 |
The Cryo-Em Structure Of Amyloid Fibrils From Abdominal Fat Of A Multiple Myeloma Patient (Case 1)- Polymorph 2.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: PROTEIN FIBRIL |
 |
The Cryo-Em Structure Of Amyloid Fibrils From Abdominal Fat Of A Multiple Myeloma Patient (Case 2).
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: PROTEIN FIBRIL |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: DNA BINDING PROTEIN/RNA/DNA Ligands: ATP, MG |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: DNA BINDING PROTEIN/RNA/DNA Ligands: ATP, MG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: PROTEIN FIBRIL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: PROTEIN FIBRIL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: PROTEIN FIBRIL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: PROTEIN FIBRIL |
 |
Cryo-Em Structure Of Mers-Cov Nsp10-Nsp14 (E191A) In Complex With T20P15 Rna, Monomeric Form
Organism: Middle east respiratory syndrome-related coronavirus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: Viral Protein/RNA Ligands: ZN, MG |
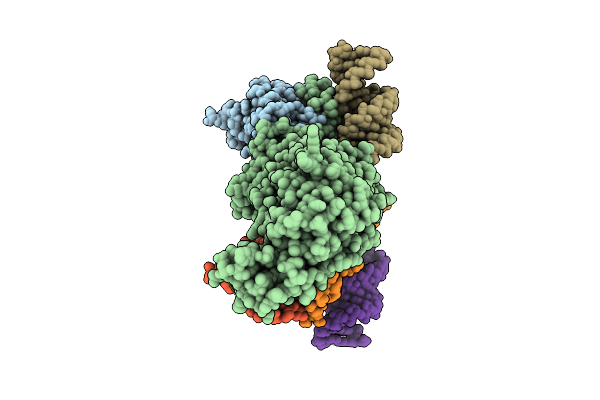 |
Cryo-Em Structure Of Mers-Cov Nsp10-Nsp14 (E191A) In Complex With T20P15 Rna, Dimeric Form
Organism: Middle east respiratory syndrome-related coronavirus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: Viral Protein/RNA Ligands: ZN, MG |
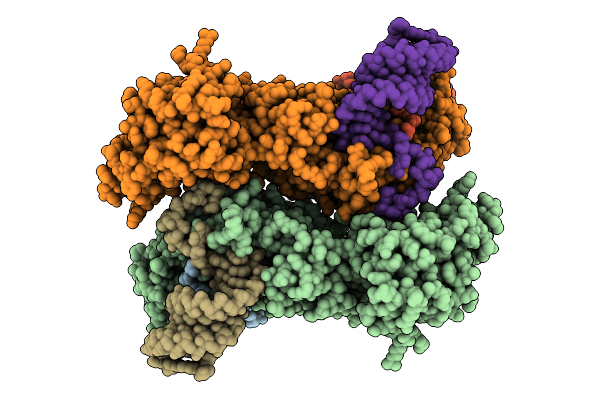 |
Cryo-Em Structure Of Mers-Cov Nsp10-Nsp14 (E191A) In Complex With T20P14-U Rna
Organism: Middle east respiratory syndrome-related coronavirus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: Viral Protein/RNA Ligands: ZN, MG |
 |
Cryo-Em Structure Of Mers-Cov Nsp10-Nsp14 (E191A) In Complex With T20P14-A Rna
Organism: Middle east respiratory syndrome-related coronavirus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RNA/VIRAL PROTEIN Ligands: ZN, MG |
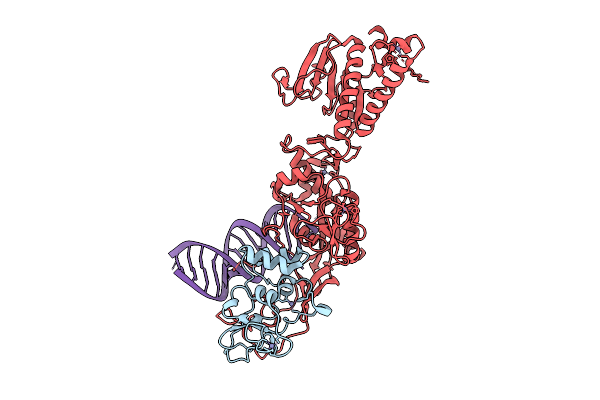 |
Cryo-Em Structure Of Mers-Cov Nsp10-Nsp14 (E191A) In Complex With T20P14-G Rna
Organism: Middle east respiratory syndrome-related coronavirus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: VIRAL PROTEIN/RNA Ligands: ZN, MG |
 |
Crystal Structure Of The Kv7.1 C-Terminal Domain In Complex With Calmodulin Disease Mutation F90L
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: METAL BINDING PROTEIN Ligands: CA |
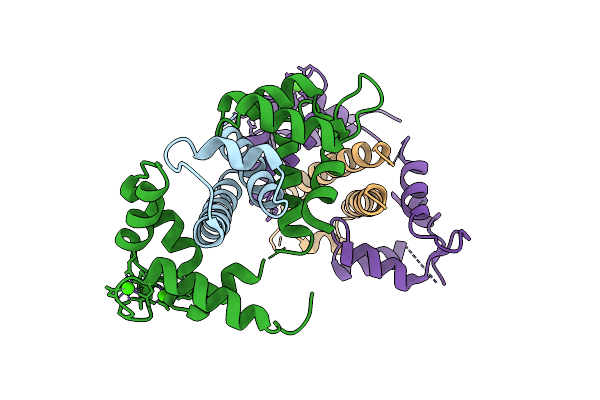 |
Crystal Structure Of The Kv7.1 C-Terminal Domain In Complex With Calmodulin Disease Mutation Q136P
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: METAL BINDING PROTEIN Ligands: CA |
 |
Crystal Structure Of The Kv7.1 C-Terminal Domain In Complex With Calmodulin Disease Mutation D130G
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: METAL BINDING PROTEIN Ligands: CA |