Search Count: 109
 |
Organism: Archangium gephyra
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2024-06-26 Classification: UNKNOWN FUNCTION Ligands: IMD, EDO, MPO, CL |
 |
Organism: Cystobacter fuscus dsm 2262
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2024-06-26 Classification: UNKNOWN FUNCTION Ligands: ACT, EDO, MPD, PG6 |
 |
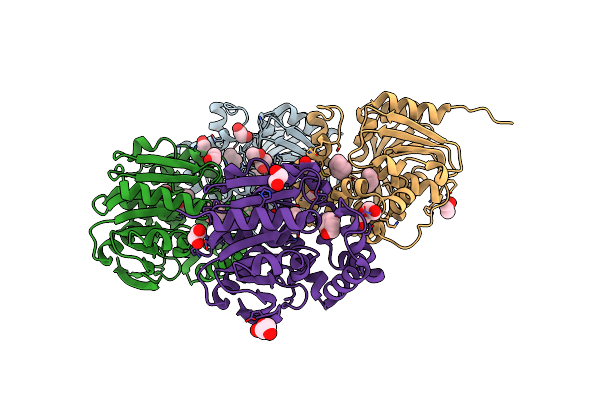
Cyclic 2,3-Diphosphoglycerate Synthetase From The Hyperthermophilic Archaeon Methanothermus Fervidus
Organism: Methanothermus fervidus dsm 2088
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2023-12-06 Classification: LIGASE Ligands: MPO, EDO, PEG, NA, CL |
 |
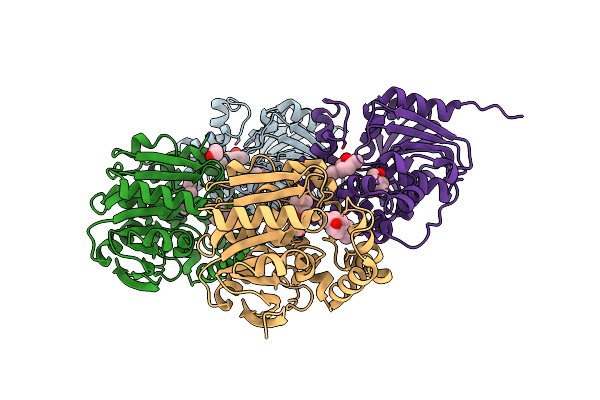
Cyclic 2,3-Diphosphoglycerate Synthetase From The Hyperthermophilic Archaeon Methanothermus Fervidus Bound To 2,3-Diphosphoglycerate And Adp.
Organism: Methanothermus fervidus dsm 2088
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2023-12-06 Classification: LIGASE Ligands: ADP, EDO, DG2, MG, PO4 |
 |
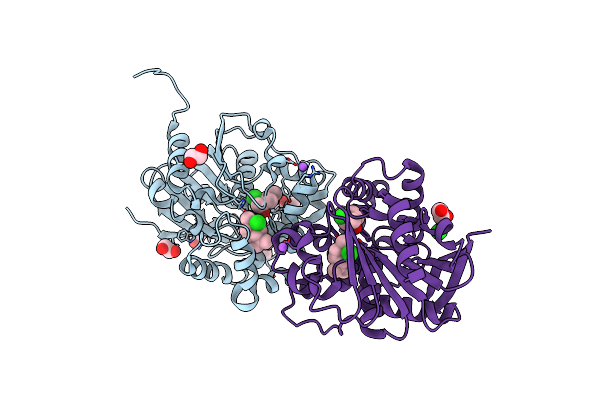
Limonene Epoxide Low Ph Soak Of Epoxide Hydrolase From Metagenomic Source Ch65
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2023-08-16 Classification: HYDROLASE Ligands: LEO, CL, EDO |
 |
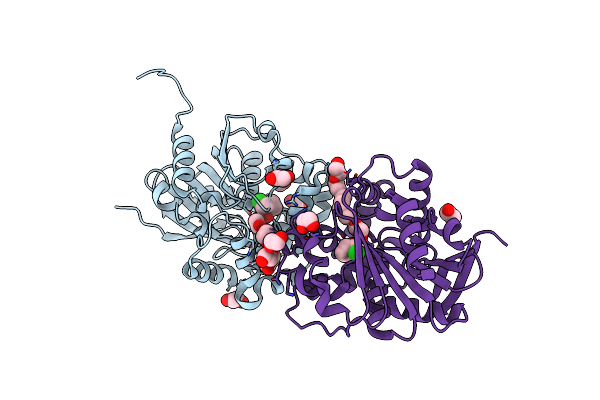
Cyclohexane Epoxide Low Ph Soak Of Epoxide Hydrolase From Metagenomic Source Ch65
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2023-08-16 Classification: HYDROLASE Ligands: 7EX, PEG, EDO |
 |
Halogenated Product Of Limonene Epoxide Turnover By Epoxide Hydrolase From Metagenomic Source Ch65
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2023-08-16 Classification: HYDROLASE Ligands: M1U, CL, EDO, NA |
 |
Cyclohexane Epoxide Soak Of Epoxide Hydrolase From Metagenomic Source Ch65 Resulting In Halogenated Compound In The Active Site
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2023-08-16 Classification: HYDROLASE Ligands: 2CH, EDO, PGE, CL, P6G |
 |
Organism: Fasciola hepatica
Method: X-RAY DIFFRACTION Release Date: 2023-01-18 Classification: ISOMERASE |
 |
Crystal Structure Of Chromobacterium Violaceum Aminotransferase In Complex With Plp-Pyruvate Adduct
Organism: Chromobacterium violaceum (strain atcc 12472 / dsm 30191 / jcm 1249 / nbrc 12614 / ncimb 9131 / nctc 9757)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-11-23 Classification: TRANSFERASE Ligands: AN7, EDO, NA, PLP, PO4, PEG |
 |
Crystal Structure Of Chromobacterium Violaceum Aminotransferase In Complex With Plp-Pyruvate Adduct
Organism: Chromobacterium violaceum (strain atcc 12472 / dsm 30191 / jcm 1249 / nbrc 12614 / ncimb 9131 / nctc 9757)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-11-23 Classification: TRANSFERASE Ligands: AN7, EDO, CL, NA |
 |
Organism: Archaeoglobus veneficus (strain dsm 11195 / snp6)
Method: X-RAY DIFFRACTION Resolution:1.27 Å Release Date: 2021-12-01 Classification: TRANSFERASE Ligands: EDO, CA, PO4, PEG, TRS |
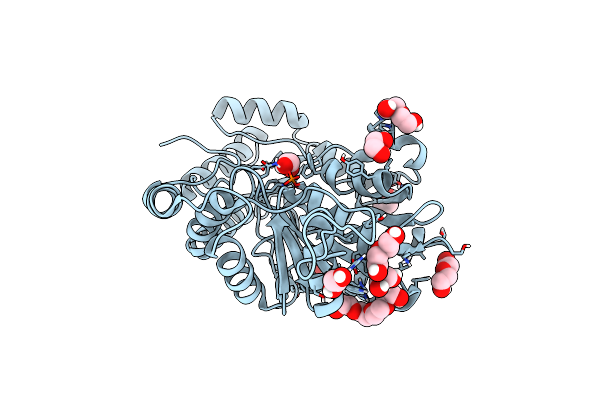 |
Sugar Transaminase From A Metagenome Collected From Troll Oil Field Production Water
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2021-12-01 Classification: TRANSFERASE Ligands: EDO, PG4, PEG |
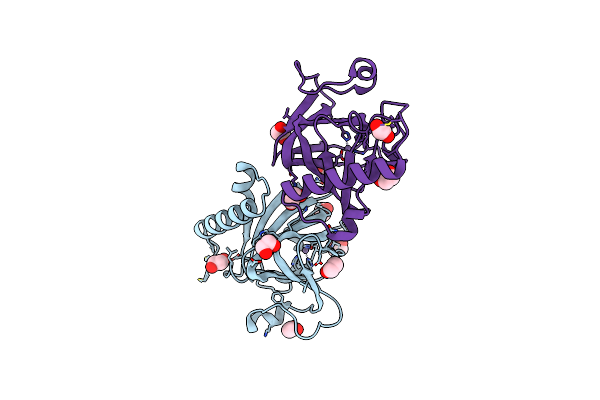 |
Organism: Thermofilum sp. ex4484_79
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2021-10-13 Classification: ISOMERASE Ligands: EDO, MN |
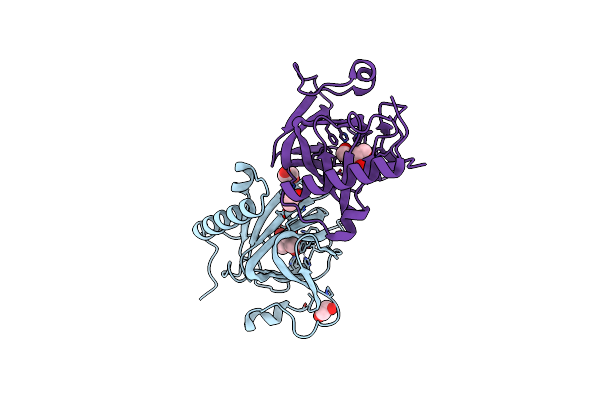 |
D-Lyxose Isomerase From The Hyperthermophilic Archaeon Thermofilum Sp Complexed With D-Fructose
Organism: Thermofilum sp. ex4484_79
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2021-10-13 Classification: ISOMERASE Ligands: FRU, EDO, MN |
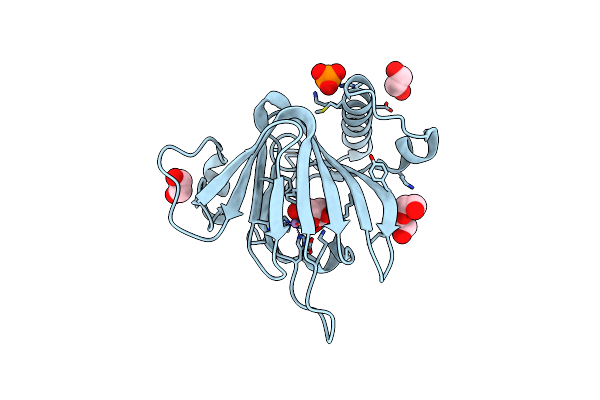 |
D-Lyxose Isomerase From The Hyperthermophilic Archaeon Thermofilum Sp Complexed With D-Mannose
Organism: Thermofilum sp. ex4484_79
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2021-10-13 Classification: ISOMERASE Ligands: BMA, EDO, PEG, MN, PO4 |
 |
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2021-05-26 Classification: HYDROLASE Ligands: EDO, ACT, SO4 |
 |
Organism: Carboxydothermus hydrogenoformans
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-11-25 Classification: TRANSFERASE Ligands: EDO, GOL, PEG |
 |
Organism: Carboxydothermus hydrogenoformans
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2020-11-25 Classification: TRANSFERASE Ligands: 8EL, CA, MLT, LMR, CL |
 |
Nad+-Dependent Fungal Formate Dehydrogenase From Chaetomium Thermophilum: A Complex With The Reduced Form Of The Cofactor Nadh And The Substrate Formate At A Secondary Site.
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719)
Method: X-RAY DIFFRACTION Resolution:1.26 Å Release Date: 2020-11-18 Classification: CYTOSOLIC PROTEIN Ligands: PEG, NAI, FMT, EDO |

