Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
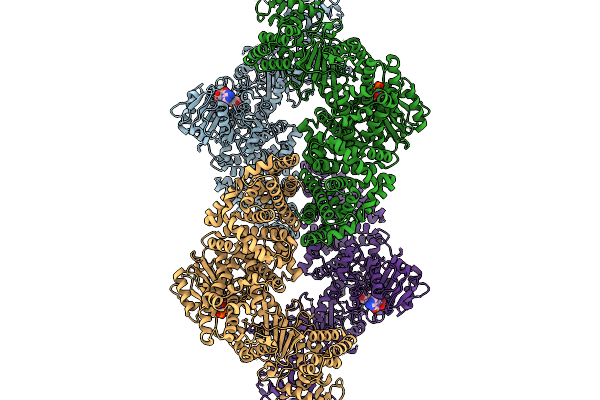 |
Cryoem Map Of The Large Glutamate Dehydrogenase Composed Of 180 Kda Subunits From Mycobacterium Smegmatis Obtained In The Presence Of Nad+ And L-Glutamate. Open Tetramer.
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: NAD |
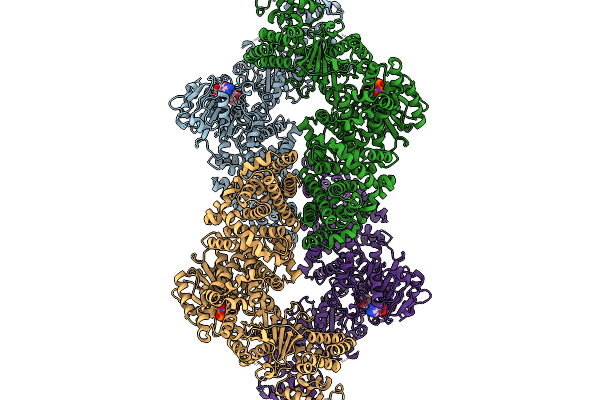 |
Cryoem Map Of The Large Glutamate Dehydrogenase Composed Of 180 Kda Subunits From Mycobacterium Smegmatis Obtained In The Presence Of Nad+ And L-Glutamate. Closed1 Tetramer.
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: NAD |
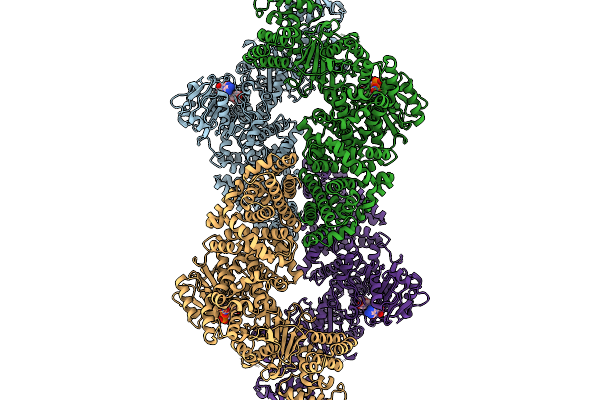 |
Cryoem Map Of The Large Glutamate Dehydrogenase Composed Of 180 Kda Subunits From Mycobacterium Smegmatis Obtained In The Presence Of Nad+ And L-Glutamate. Closed2 Tetramer
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Cryoem Map Of The Large Glutamate Dehydrogenase Composed Of 180 Kda Subunits From Mycobacterium Smegmatis Obtained In The Presence Of Nad+ And L-Glutamate. Empty Monomer.
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: OXIDOREDUCTASE |
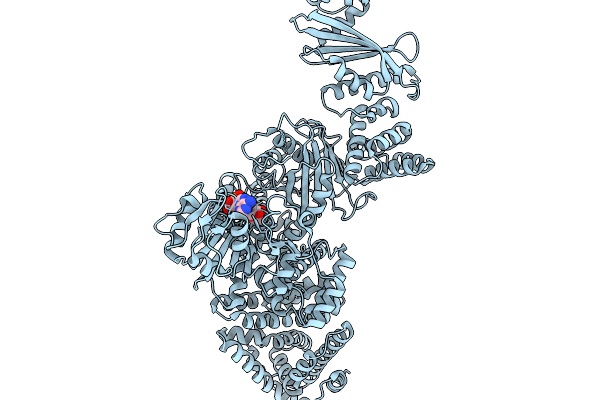 |
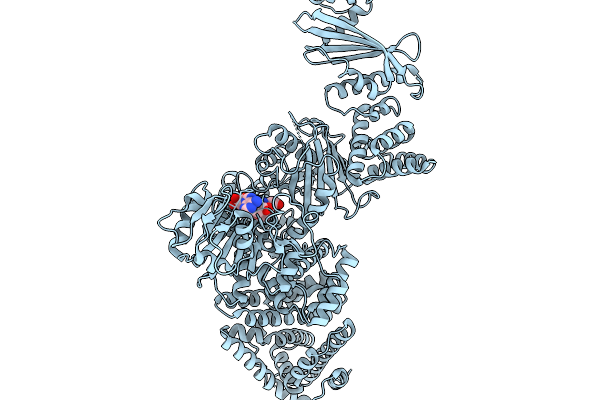
Cryoem Map Of The Large Glutamate Dehydrogenase Composed Of 180 Kda Subunits From Mycobacterium Smegmatis Obtained In The Presence Of Nad+ And L-Glutamate. Cofactor-Monomer.
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
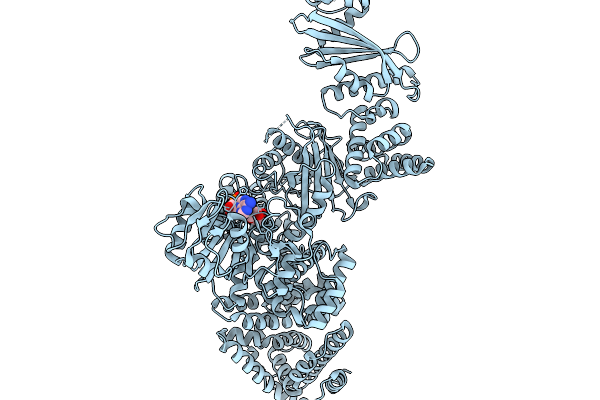
Cryoem Map Of The Large Glutamate Dehydrogenase Composed Of 180 Kda Subunits From Mycobacterium Smegmatis Obtained In The Presence Of Nad+ And L-Glutamate. Cofactor/Ligand-Monomer
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: GLU, NAD |
 |
Cryoem Map Of The Large Glutamate Dehydrogenase Composed Of 180 Kda Subunits From Mycobacterium Smegmatis Obtained In The Presence Of Nad+ And L-Glutamate. Total-Monomer
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: NAD |
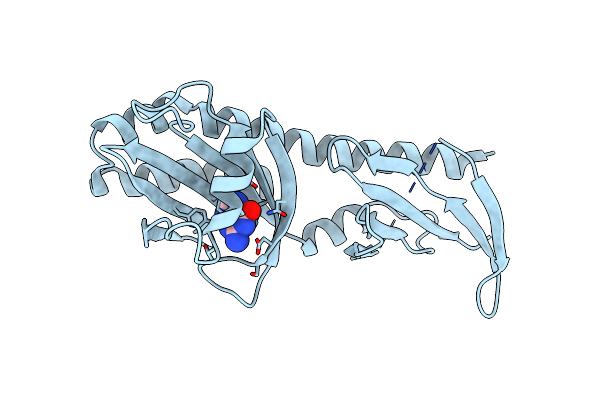 |
Crystal Structure Of The Ligand Binding Domain Of The Halomonas Titanicae Chemoreceptor Htc10 In Complex With Guanine
Organism: Halomonas titanicae
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2024-11-20 Classification: SIGNALING PROTEIN Ligands: GUN |
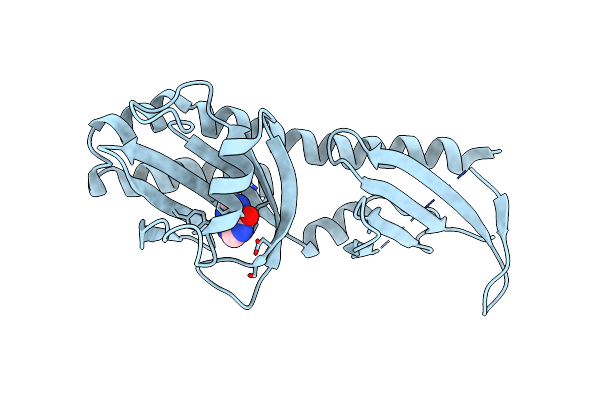 |
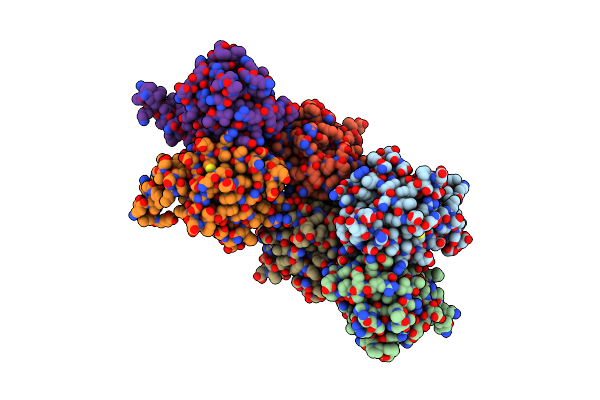
Crystal Structure Of The Ligand Binding Domain Of The Halomonas Titanicae Chemoreceptor Htc10 In Complex With Hypoxanthine
Organism: Halomonas titanicae
Method: X-RAY DIFFRACTION Resolution:3.63 Å Release Date: 2024-11-20 Classification: SIGNALING PROTEIN Ligands: HPA |
 |
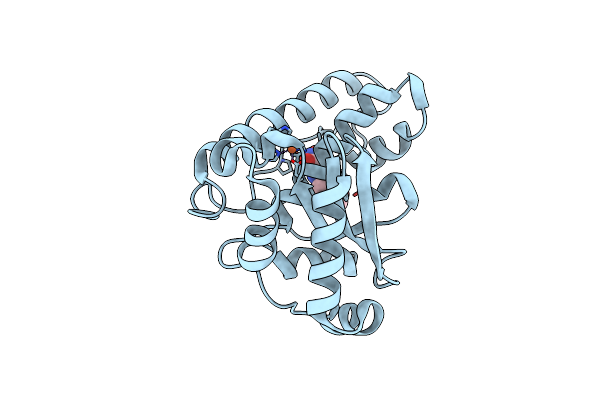
High-Resolution Crystal Structure Of The Ligand Binding Domain Of The Halomonas Titanicae Chemoreceptor Htc10 In Complex With Guanine
Organism: Halomonas titanicae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-11-20 Classification: SIGNALING PROTEIN Ligands: GUN |
 |
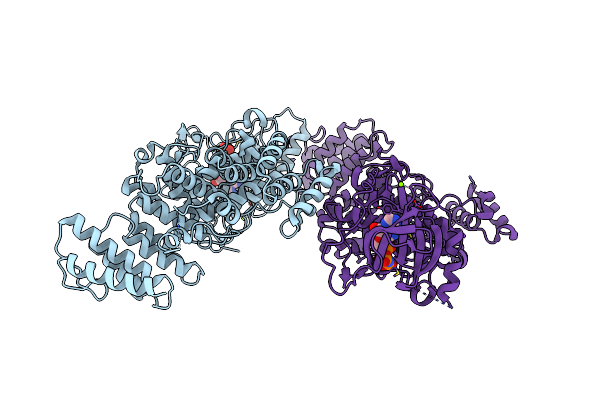
Crystal Structure Of The Iron Superoxide Dismutase From Acinetobacter Sp. Ver3
Organism: Acinetobacter sp. ver3
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2022-03-16 Classification: OXIDOREDUCTASE Ligands: FMN, FE |
 |
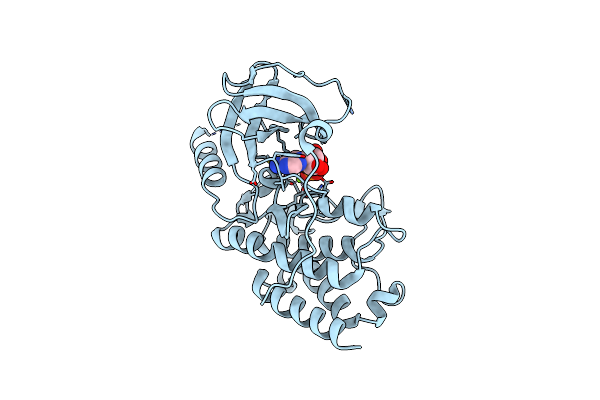
Crystal Structure Of The S/T Protein Kinase Pkng From Corynebacterium Glutamicum In Complex With Amp-Pnp
Organism: Corynebacterium glutamicum (strain atcc 13032 / dsm 20300 / bcrc 11384 / jcm 1318 / lmg 3730 / ncimb 10025)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2021-10-13 Classification: TRANSFERASE Ligands: MAP, MG |
 |
Crystal Structure Of The S/T Protein Kinase Pkng From Corynebacterium Glutamicum (Residues 130-433) In Complex With Amp-Pnp, Isoform 1
Organism: Corynebacterium glutamicum (strain atcc 13032 / dsm 20300 / bcrc 11384 / jcm 1318 / lmg 3730 / ncimb 10025)
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2021-10-13 Classification: TRANSFERASE Ligands: MG, MAP |
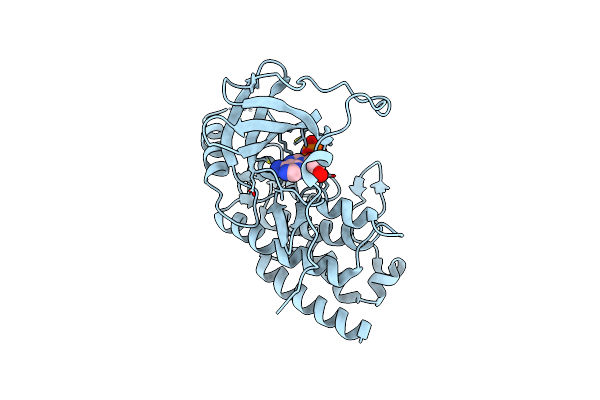 |
Crystal Structure Of The S/T Protein Kinase Pkng From Corynebacterium Glutamicum (Residues 130-433) In Complex With Amp-Pnp, Isoform 2
Organism: Corynebacterium glutamicum (strain atcc 13032 / dsm 20300 / bcrc 11384 / jcm 1318 / lmg 3730 / ncimb 10025)
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2021-10-13 Classification: TRANSFERASE Ligands: MG, MAP |
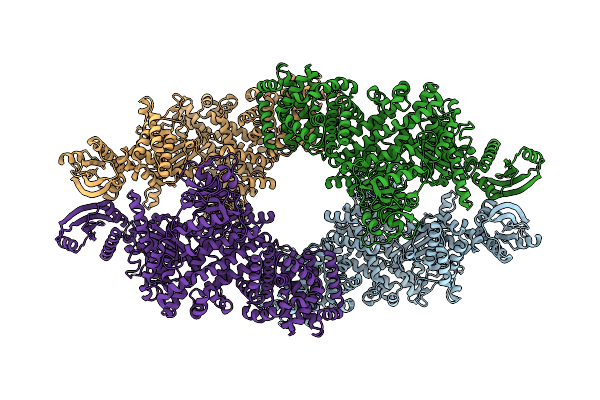 |
Cryo-Em Map Of The Large Glutamate Dehydrogenase Composed Of 180 Kda Subunits From Mycobacterium Smegmatis (Open Conformation)
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2021-06-09 Classification: OXIDOREDUCTASE |
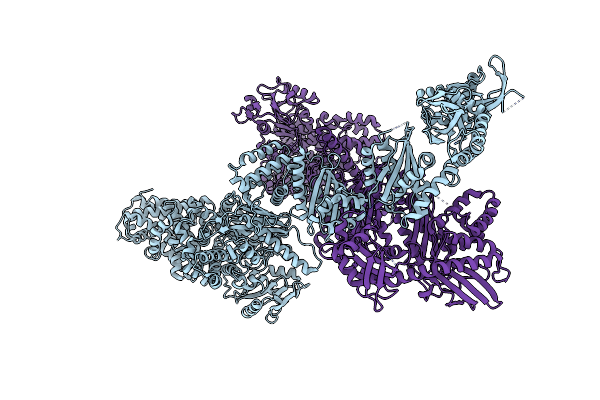 |
Crystal Structure Of The Large Glutamate Dehydrogenase Composed Of 180 Kda Subunits From Mycobacterium Smegmatis
Organism: Mycolicibacterium smegmatis
Method: X-RAY DIFFRACTION Resolution:6.27 Å Release Date: 2021-06-09 Classification: OXIDOREDUCTASE |
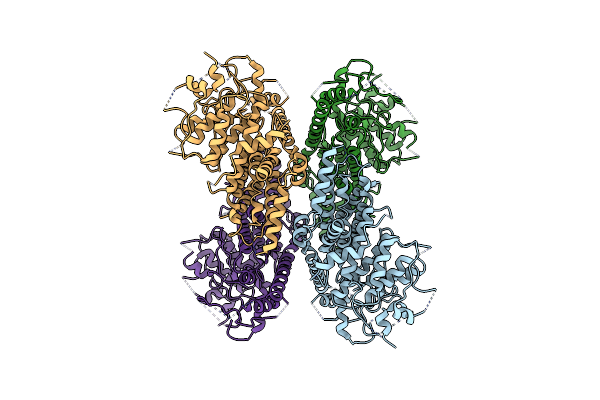 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2020-11-18 Classification: CYTOSOLIC PROTEIN |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2020-09-23 Classification: MEMBRANE PROTEIN |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2019-11-20 Classification: ELECTRON TRANSPORT, OXIDOREDUCTASE Ligands: CUA |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2019-11-20 Classification: ELECTRON TRANSPORT, OXIDOREDUCTASE Ligands: CUA, GOL, SO4 |