Search Count: 96
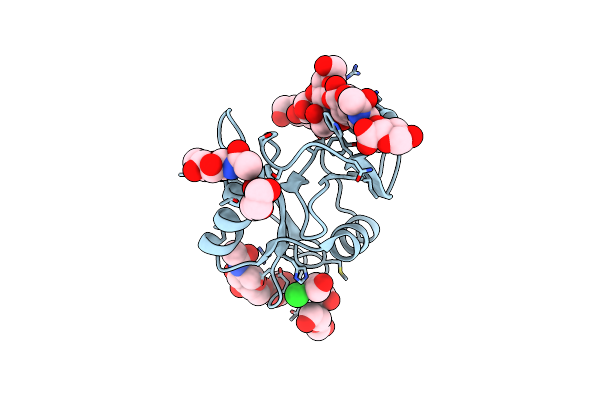 |
Structure Of The Murine Lyve-1 (Lymphatic Vessel Endothelial Receptor-1) Hyaluronan Binding Domain Bound With Octasaccharide Hyaluronan.
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2025-02-12 Classification: CELL ADHESION Ligands: NAG, GOL, PEG, CL |
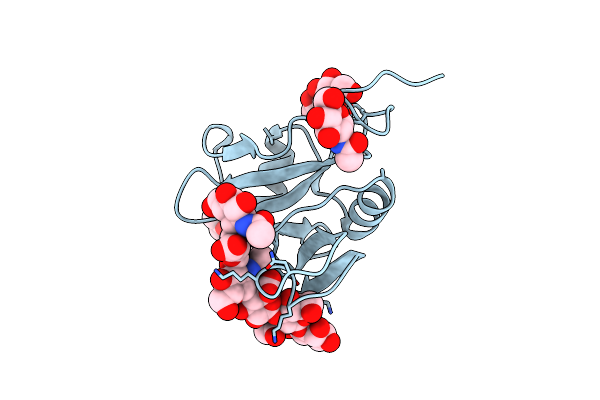 |
Structure Of The Human Lyve-1 (Lymphatic Vessel Endothelial Receptor-1) Hyaluronan Binding Domain Bound With Decasaccharide Hyaluronan.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.32 Å Release Date: 2024-11-13 Classification: CELL ADHESION Ligands: NAG |
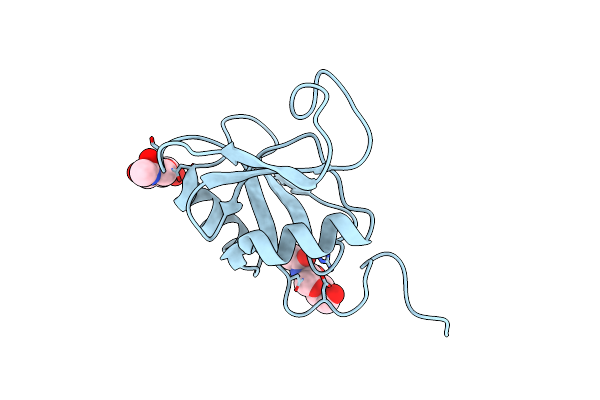 |
Structure Of The Murine Lyve-1 (Lymphatic Vessel Endothelial Receptor-1) Hyaluronan Binding Domain In An Unliganded State
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2024-10-16 Classification: CELL ADHESION Ligands: NAG |
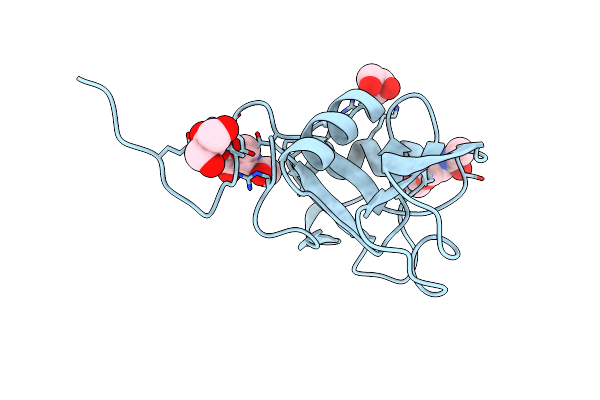 |
Structure Of The Human Lyve-1 (Lymphatic Vessel Endothelial Receptor-1) Hyaluronan Binding Domain In An Unliganded State
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2024-07-31 Classification: CELL ADHESION Ligands: NAG, GOL |
 |
Elongating E. Coli 70S Ribosome Containing Deacylated Trna(Imet) In The P-Site And Aaa Mrna Codon With Cognate Dipeptidyl-Trna(Lys) In The A-Site
Organism: Synthetic construct, Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2023-08-16 Classification: TRANSLATION |
 |
Elongating E. Coli 70S Ribosome Containing Acylated Trna(Imet) In The P-Site And Aam6A Mrna Codon In The A-Site After Uncompleted Di-Peptide Formation
Organism: Synthetic construct, Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2023-08-16 Classification: TRANSLATION Ligands: FME |
 |
Elongating E. Coli 70S Ribosome Containing Acylated Trna(Imet) In The P-Site And Aaa Mrna Codon In The A-Site After Uncompleted Di-Peptide Formation
Organism: Synthetic construct, Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2023-08-16 Classification: TRANSLATION Ligands: FME |
 |
Elongating E. Coli 70S Ribosome Containing Deacylated Trna(Imet) In The P-Site And Aam6A Mrna Codon With Cognate Dipeptidyl-Trna(Lys) In The A-Site
Organism: Synthetic construct, Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2023-08-16 Classification: TRANSLATION |
 |
Elongating E. Coli 70S Ribosome Containing Deacylated Trna(Imet) In The P-Site And Am6Aa Mrna Codon With Cognate Dipeptidyl-Trna(Lys) In The A-Site
Organism: Synthetic construct, Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2023-08-16 Classification: TRANSLATION |
 |
Elongating E. Coli 70S Ribosome Containing Acylated Trna(Imet) In The P-Site And Am6Aa Mrna Codon In The A-Site After Uncompleted Di-Peptide Formation
Organism: Synthetic construct, Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2023-08-16 Classification: TRANSLATION Ligands: FME |
 |
Elongating E. Coli 70S Ribosome Containing Deacylated Trna(Imet) In The P-Site And M6Aaa Mrna Codon With Cognate Dipeptidyl-Trna(Lys) In The A-Site
Organism: Synthetic construct, Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2023-08-16 Classification: TRANSLATION |
 |
Elongating E. Coli 70S Ribosome Containing Acylated Trna(Imet) In The P-Site And M6Aaa Mrna Codon In The A-Site After Uncompleted Di-Peptide Formation
Organism: Synthetic construct, Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2023-08-16 Classification: TRANSLATION Ligands: FME |
 |
Initiation Complex Of The E. Coli 70S Ribosome With Mrna Containing Aaa Codon In The A-Site.
Organism: Synthetic construct, Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2023-08-16 Classification: TRANSLATION Ligands: FME |
 |
Initiation Complex Of The E. Coli 70S Ribosome With Mrna Containing Aam6A Codon In The A-Site
Organism: Synthetic construct, Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2023-08-16 Classification: TRANSLATION Ligands: FME |
 |
An Irreversible, Promiscuous And Highly Thermostable Claisen-Condensation Biocatalyst Drives The Synthesis Of Substituted Pyrroles
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-09-21 Classification: SYNTHASE Ligands: PLP, MPD, SO4 |
 |
An Irreversible, Promiscuous And Highly Thermostable Claisen-Condensation Biocatalyst Drives The Synthesis Of Substituted Pyrroles
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-09-21 Classification: SYNTHASE Ligands: PLP, NA |
 |
An Irreversible, Promiscuous And Highly Thermostable Claisen-Condensation Biocatalyst Drives The Synthesis Of Substituted Pyrroles
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-09-21 Classification: SYNTHASE Ligands: PLP |
 |
Crystal Structure Of Human Pi3Ka (P110A Subunit) With Mmv085400 Bound To The Active Site Determined At 2.9 Angstroms Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2022-05-04 Classification: TRANSFERASE/INHIBITOR Ligands: ZHY |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-12-22 Classification: DNA Ligands: BA |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.32 Å Release Date: 2021-10-06 Classification: DNA Ligands: SR, NA, NCO |

