Search Count: 8
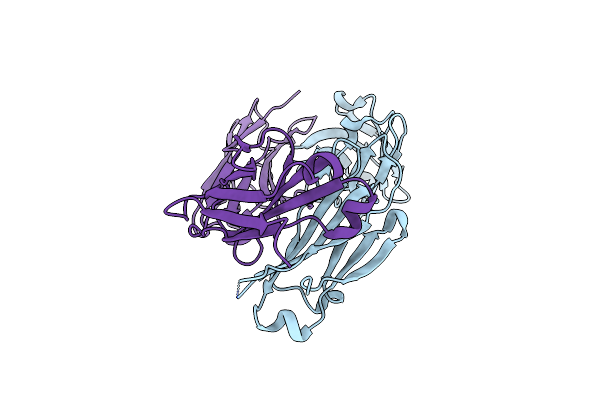 |
Ablift: Antibody Stability And Affinity Optimization By Computational Design Of The Variable Light-Heavy Chain Interface
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2019-05-01 Classification: IMMUNE SYSTEM |
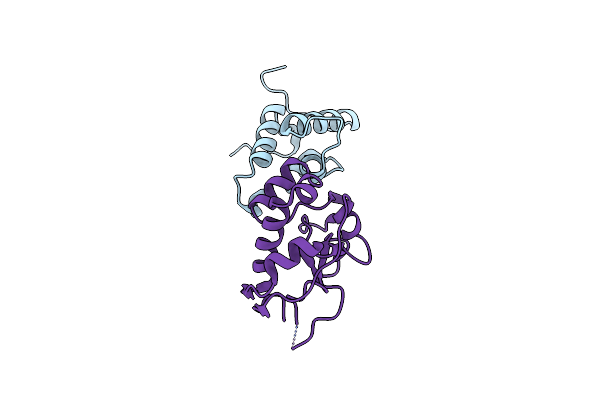 |
Crystal Structure Of A Computationally Designed Colicin Endonuclease And Immunity Pair Coledes7/Imdes7
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2019-01-30 Classification: HYDROLASE |
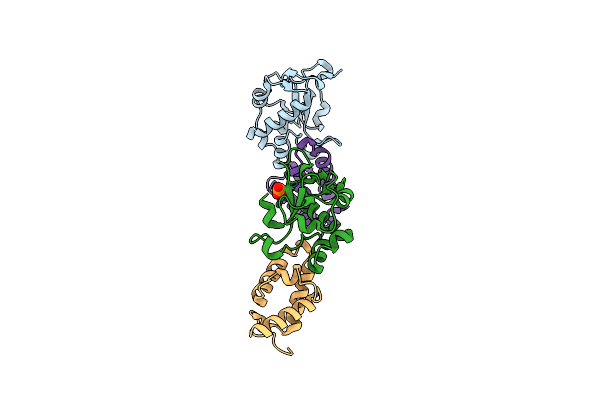 |
Crystal Structure Of A Computationally Designed Colicin Endonuclease And Immunity Pair Coledes3/Imdes3
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2019-01-30 Classification: IMMUNE SYSTEM Ligands: PO4 |
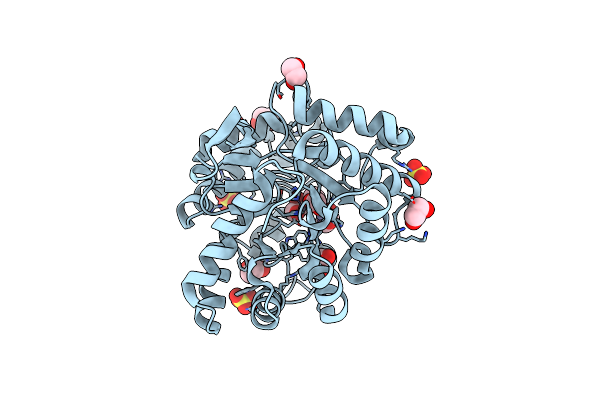 |
Repertoires Of Functionally Diverse Enzymes Through Computational Design At Epistatic Active-Site Positions
Organism: Brevundimonas diminuta
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2018-10-24 Classification: HYDROLASE Ligands: FMT, ZN, EDO, SO4 |
 |
Repertoires Of Functionally Diverse Enzymes Through Computational Design At Epistatic Active-Site Positions
Organism: Brevundimonas diminuta
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-10-24 Classification: HYDROLASE Ligands: FMT, ZN, TRS |
 |
Repertoires Of Functionally Diverse Enzymes Through Computational Design At Epistatic Active-Site Positions
Organism: Brevundimonas diminuta
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2018-10-24 Classification: HYDROLASE Ligands: FMT, ZN, EDO, CAC |
 |
Highly Active Enzymes By Automated Modular Backbone Assembly And Sequence Design
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2018-07-25 Classification: Artificial enzyme |
 |
Highly Active Enzymes By Automated Modular Backbone Assembly And Sequence Design
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2018-07-25 Classification: Artificial enzyme |

