Search Count: 71
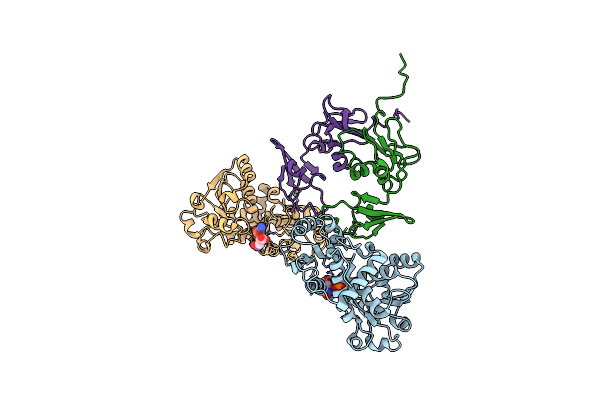 |
T-State Active Site Of Aspartate Transcarbamylase:Crystal Structure Of The Carbamyl Phosphate And L-Alanosine Ligated Enzyme
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-01-24 Classification: TRANSFERASE Ligands: CP, AL0, ZN |
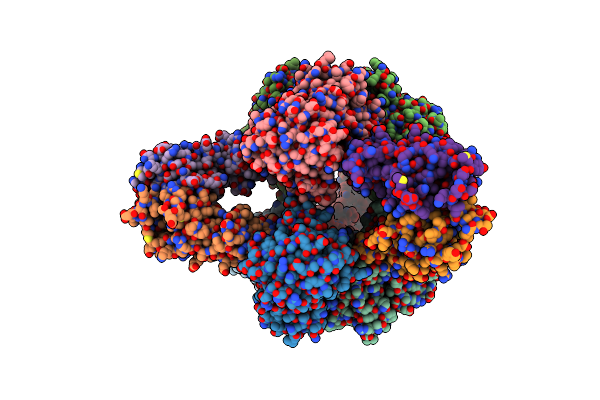 |
Aspartate Transcarbamylase (Atcase) Of Escherichia Coli: A New Crystalline R State Bound To Pala, Or To Product Analogues Phosphate And Citrate
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2004-06-08 Classification: TRANSFERASE Ligands: PAL, ZN |
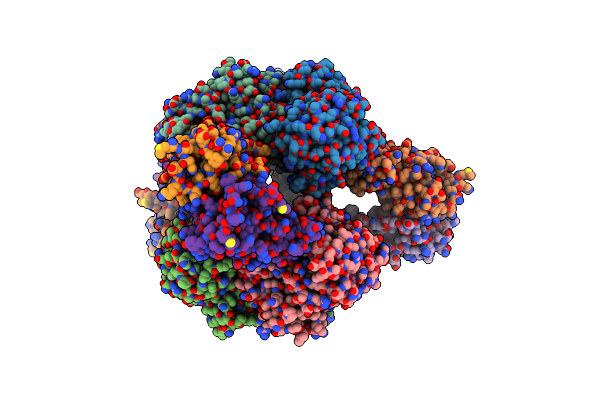 |
Aspartate Transcarbamylase (Atcase) Of Escherichia Coli: A New Crystalline R State Bound To Pala, Or To Product Analogues Phosphate And Citrate
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2004-06-08 Classification: TRANSFERASE Ligands: FLC, PO4, ZN |
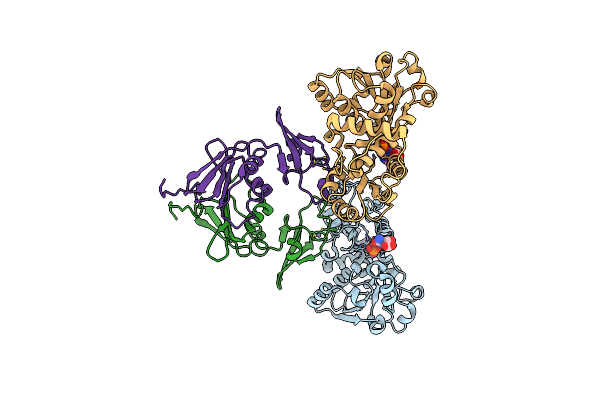 |
Products In The T State Of Aspartate Transcarbamylase: Crystal Structure Of The Phosphate And N-Carbamyl-L-Aspartate Ligated Enzyme
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2004-06-08 Classification: TRANSFERASE Ligands: PO4, NCD, ZN |
 |
Crystal Structure Of The Tyrosine-Regulated 3-Deoxy-D-Arabino-Heptulosonate-7-Phosphate Synthase From Saccharomyces Cerevisiae Complexed With Phosphoenolpyruvate
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2003-01-14 Classification: LYASE Ligands: PEP |
 |
Aspartate Transcarbamoylase Complexed With N-Phosphonacetyl-L-Aspartate (Pala)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2000-01-28 Classification: TRANSFERASE Ligands: PAL, ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1998-06-17 Classification: INTERFERON Ligands: ZN |
 |
Structure Of Yeast Chorismate Mutase With Bound Trp And An Endooxabicyclic Inhibitor
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 1998-01-14 Classification: COMPLEX (ISOMERASE/PEPTIDE) Ligands: TRP, TSA |
 |
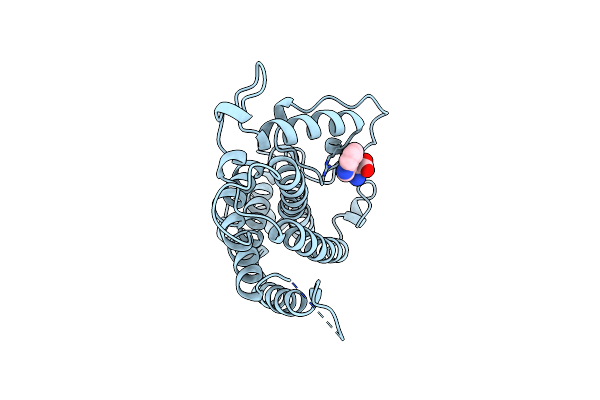
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 1998-01-14 Classification: COMPLEX (ISOMERASE/PEPTIDE) Ligands: TYR, TSA |
 |
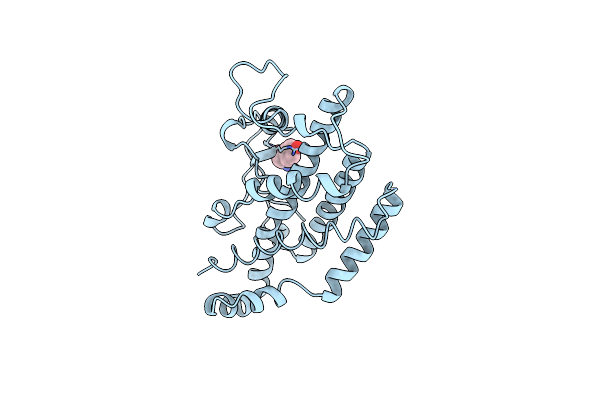
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1998-01-14 Classification: COMPLEX (ISOMERASE/PEPTIDE) Ligands: TRP |
 |
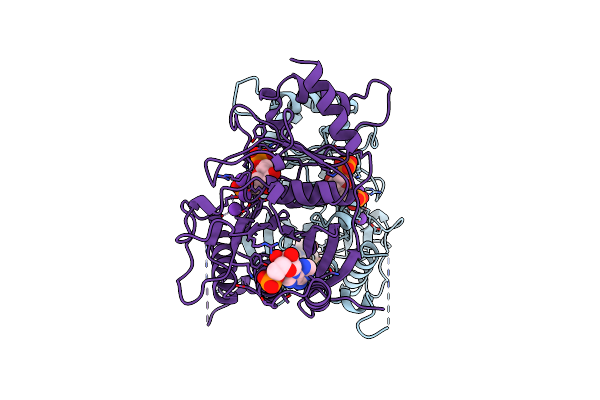
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 1996-12-23 Classification: COMPLEX (ISOMERASE/PEPTIDE) Ligands: TYR |
 |
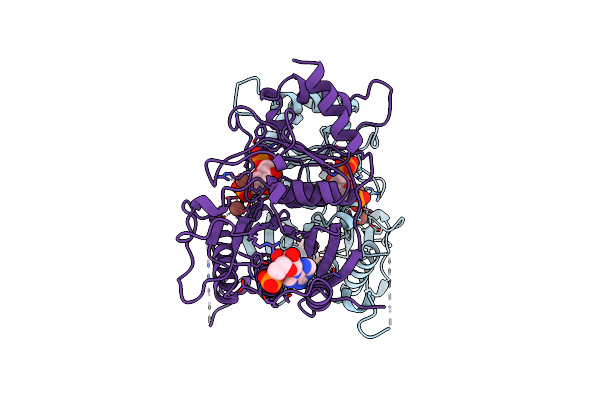
Fructose-1,6-Bisphosphatase (D-Fructose-1,6-Bisphosphate 1-Phosphohydrolase) Complexed With Amp, 2,5-Anhydro-D-Glucitol-1,6-Bisphosphate And Potassium Ions (100 Mm)
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 1996-06-20 Classification: HYDROLASE (PHOSPHORIC MONOESTER) Ligands: K, AMP, AHG |
 |
Fructose-1,6-Bisphosphatase (D-Fructose-1,6-Bisphosphate 1-Phosphohydrolase) Complexed With Amp, 2,5-Anhydro-D-Glucitol-1,6-Bisphosphate, Thallium (10 Mm) And Lithium Ions (10 Mm)
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1996-06-20 Classification: HYDROLASE (PHOSPHORIC MONOESTER) Ligands: TL, AMP, AHG |
 |
Fructose-1,6-Bisphosphatase (D-Fructose-1,6-Bisphosphate 1-Phosphohydrolase) Complexed With Thallium Ions (10 Mm)
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 1996-06-20 Classification: HYDROLASE (PHOSPHORIC MONOESTER) Ligands: TL |
 |
Fructose-1,6-Bisphosphatase (D-Fructose-1,6-Bisphosphate 1-Phosphohydrolase) Complexed With Amp, 2,5-Anhydro-D-Glucitol-1,6-Bisphosphate And Thallium Ions (10 Mm)
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 1996-06-20 Classification: HYDROLASE (PHOSPHORIC MONOESTER) Ligands: TL, AMP, AHG |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 1995-10-15 Classification: HYDROLASE (ALPHA-AMINOACYLPEPTIDE) Ligands: ZN, CO3, MRD |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1995-10-15 Classification: HYDROLASE (ALPHA-AMINOACYLPEPTIDE) Ligands: ZN, LEU, MRD |
 |
The Crystal Structure Of Allosteric Chorismate Mutase At 2.2 Angstroms Resolution
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1995-09-15 Classification: ISOMERASE Ligands: TRP |
 |
Organism: Haemophilus influenzae biotype aegyptius
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 1995-09-15 Classification: TRANSFERASE/DNA Ligands: CA |
 |
Bovine Lens Leucine Aminopeptidase Complexed With L-Leucine Phosphonic Acid
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 1995-07-31 Classification: HYDROLASE (ALPHA-AMINOACYLPEPTIDE) Ligands: ZN, PLU, MRD |

