Search Count: 24
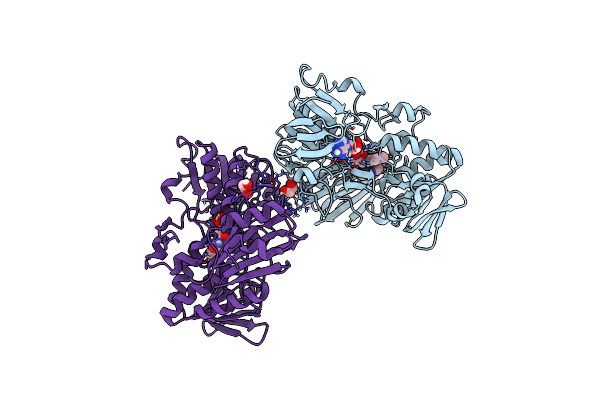 |
Crystal Structure Of Human Apoptosis-Inducing Factor (Aif) W196A Mutant Complexed With 6-Fluoroquinolin-4-Amine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2023-11-08 Classification: OXIDOREDUCTASE Ligands: FAD, QDI, EDO, CL |
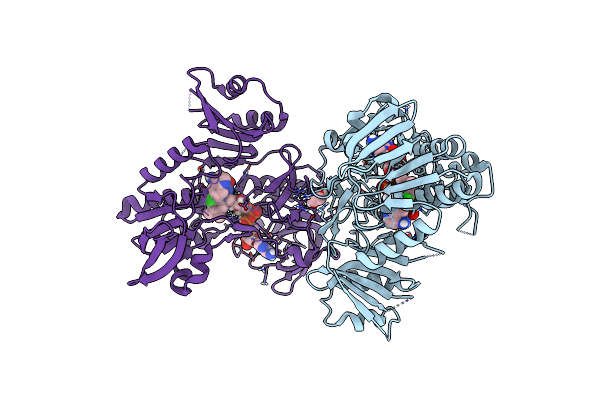 |
Crystal Structure Of Human Apoptosis-Inducing Factor (Aif) W196A Mutant Complexed With 6-Chloroquinolin-4-Amine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2023-11-08 Classification: OXIDOREDUCTASE Ligands: FAD, QD1, EDO |
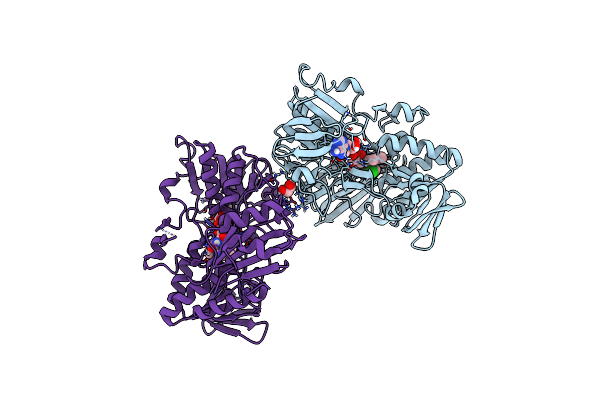 |
Crystal Structure Of Human Apoptosis-Inducing Factor (Aif) W196A Mutant Complexed With 7-Chloroquinolin-4-Amine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2023-11-08 Classification: OXIDOREDUCTASE Ligands: FAD, QBC, EDO |
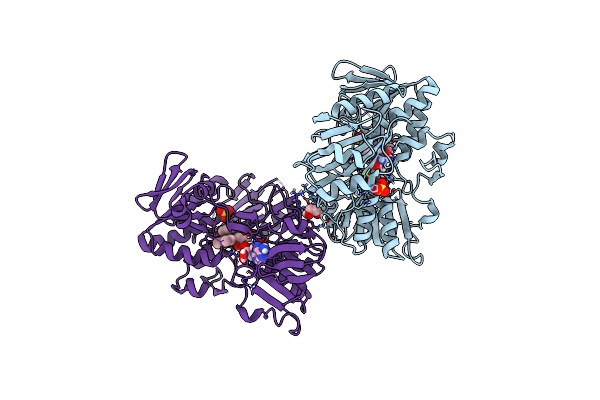 |
Crystal Structure Of Human Apoptosis-Inducing Factor (Aif) W196A Mutant Complexed With Quinolin-4-Amine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2023-11-08 Classification: OXIDOREDUCTASE Ligands: 1LM, EDO, FAD, SO4 |
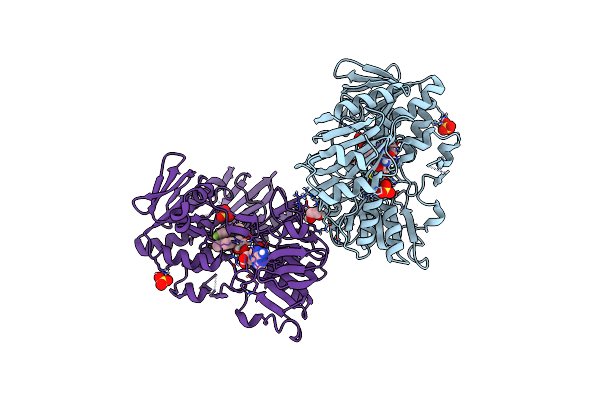 |
Crystal Structure Of Human Apoptosis-Inducing Factor (Aif) Complexed With 6-Fluoro-2-Methylquinolin-4-Amine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-11-08 Classification: OXIDOREDUCTASE Ligands: EDO, FAD, QEF, SO4 |
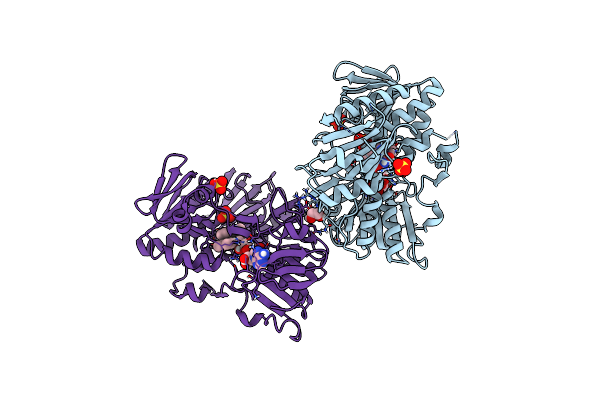 |
Crystal Structure Of Human Apoptosis-Inducing Factor (Aif) Complexed With 8-Fluoro-2-Methylquinolin-4-Amine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-11-08 Classification: OXIDOREDUCTASE Ligands: QC6, EDO, FAD, SO4 |
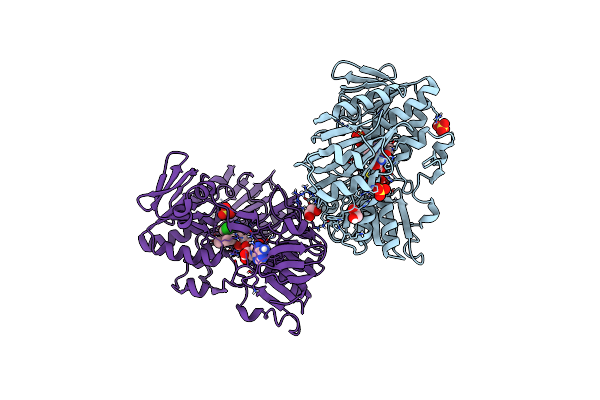 |
Crystal Structure Of Human Apoptosis-Inducing Factor (Aif) Complexed With 7-Chloroquinolin-4-Amine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2023-11-08 Classification: OXIDOREDUCTASE Ligands: QBC, FAD, EDO, SO4 |
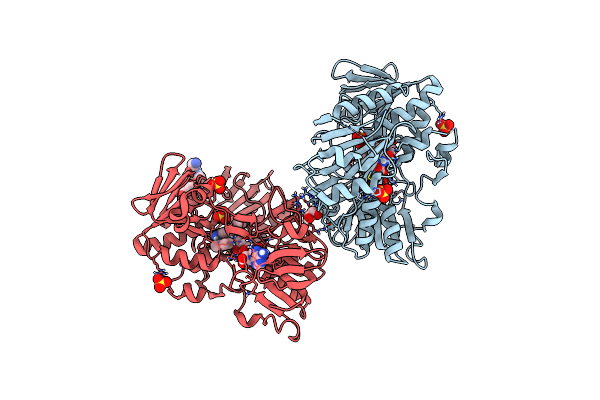 |
Crystal Structure Of Human Apoptosis-Inducing Factor (Aif) Complexed With 8-Methoxyquinolin-4-Amine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2023-11-08 Classification: OXIDOREDUCTASE Ligands: QF6, EDO, FAD, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2017-09-27 Classification: LYASE Ligands: MG, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2016-07-20 Classification: LYASE/LYASE INHIBITOR Ligands: 4NG, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2016-04-13 Classification: LYASE/LYASE INHIBITOR Ligands: MG, PAH |
 |
Pkg Ii'S Carboxyl Terminal Cyclic Nucleotide Binding Domain (Cnb-B) In A Complex With Cgmp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2016-01-20 Classification: TRANSFERASE Ligands: 35G, CA, ACT, NA |
 |
Pkg Ii'S Amino Terminal Cyclic Nucleotide Binding Domain (Cnb-A) In A Complex With Camp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2016-01-20 Classification: PROTEIN BINDING Ligands: CD, CMP, CO, CA, EDO, NA |
 |
Pkg Ii'S Amino Terminal Cyclic Nucleotide Binding Domain (Cnb-A) In A Complex With Cgmp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-01-20 Classification: TRANSFERASE Ligands: MLA, PCG, NA |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2013-12-11 Classification: RNA BINDING PROTEIN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2013-12-11 Classification: RNA BINDING PROTEIN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2013-12-11 Classification: RNA BINDING PROTEIN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2013-12-11 Classification: RNA BINDING PROTEIN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2012-10-03 Classification: TRANSFERASE Ligands: CL, PO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2012-10-03 Classification: TRANSFERASE Ligands: CL, PO4 |

