Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
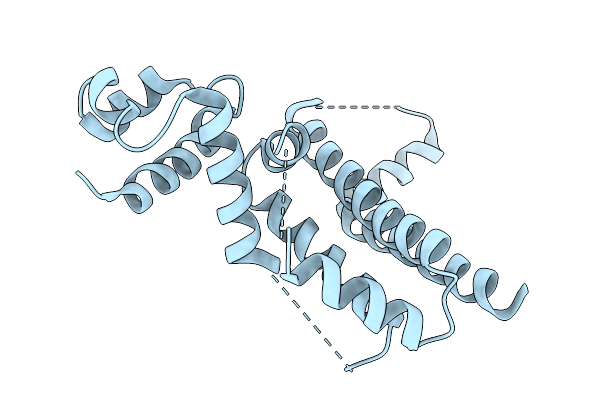 |
Crystal Structure Of The Engineered Sulfonylurea Repressor Esr (L7-D1), Apo Form
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: DNA BINDING PROTEIN |
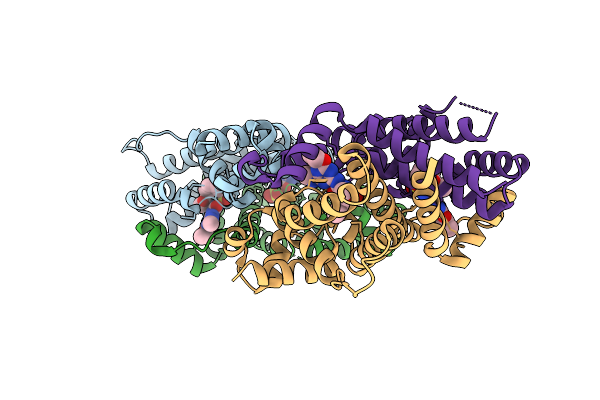 |
Crystal Structure Of The Engineered Sulfonylurea Repressor Esr (L11-C6), Bound To Ethametsulfuron-Methyl
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: DNA BINDING PROTEIN Ligands: RXF |
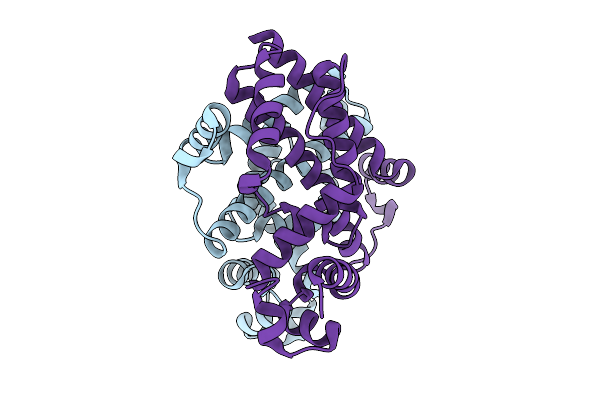 |
Crystal Structure Of The Engineered Sulfonylurea Repressor Csr (L4.2-20), Apo Form
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: DNA BINDING PROTEIN |
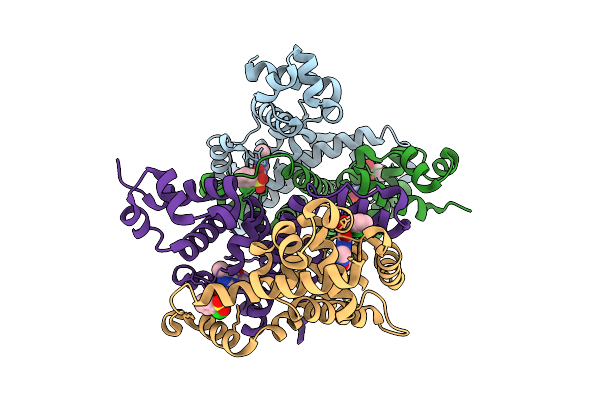 |
Crystal Structure Of The Engineered Sulfonylurea Repressor Csr (L4.2-20), Bound To Chlorsulfuron
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: DNA BINDING PROTEIN Ligands: 1CS |
 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: HYDROLASE |
 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: HYDROLASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.26 Å Release Date: 2023-11-22 Classification: PEPTIDE BINDING PROTEIN Ligands: ZJ9, EDO, DMS, PO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-11-22 Classification: PEPTIDE BINDING PROTEIN Ligands: EDO, DMS, ZJF, SO4 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-06-14 Classification: LIGASE Ligands: ZN |
 |
 |
Crystal Structure Of Human Yeats4 In Complex With Pfizer Small Molecule Compound 3B
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2023-01-11 Classification: PROTEIN BINDING Ligands: SJI |
 |
Cryo-Em Structure Of Rcmv-E E27 Bound To Human Ddb1 (Deltabpb) And Rat Stat2 Ccd
Organism: Homo sapiens, Murid betaherpesvirus 8, Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2022-11-09 Classification: VIRUS Ligands: ZN |
 |
Cryo-Em Structure Of Rcmv-E E27 Bound To Human Ddb1 (Deltabpb) And Full-Length Rat Stat2
Organism: Homo sapiens, Murid betaherpesvirus 8, Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2022-11-09 Classification: VIRUS Ligands: ZN |
 |
Crystal Structure Of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 Complexed With Nn-390
Organism: Danio rerio
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-11-02 Classification: METAL BINDING PROTEIN/INHIBITOR Ligands: O2L, ZN, K |
 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2022-08-10 Classification: HYDROLASE |
 |
Cryo-Em Structure Of The 20S Alpha 3 Deletion Proteasome Core Particle In Complex With Fub1
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2022-08-10 Classification: HYDROLASE |
 |
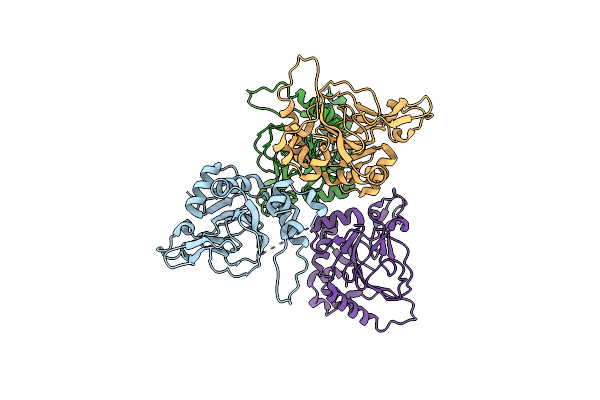
Heterodimer Structure Of Fe(Ii)/(Alpha)Ketoglutarate-Dependent Dioxygenase Tlxij
Organism: Talaromyces purpureogenus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-04-20 Classification: OXIDOREDUCTASE Ligands: OGA, FE |
 |
Organism: Talaromyces purpureogenus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-04-20 Classification: OXIDOREDUCTASE |
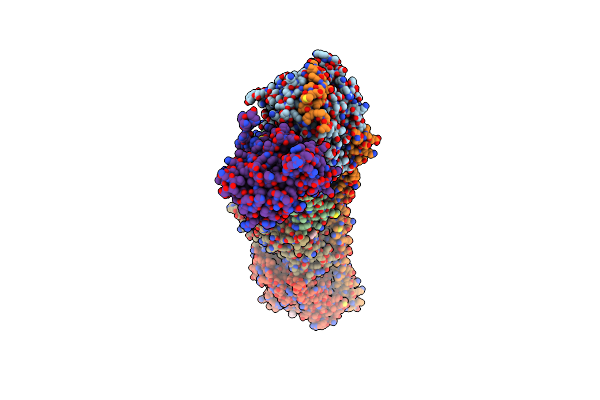 |
Organism: Mus musculus, Gallus gallus, Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2021-08-04 Classification: CELL CYCLE Ligands: GTP, MG, CA, GDP, MES, G70, ACP |
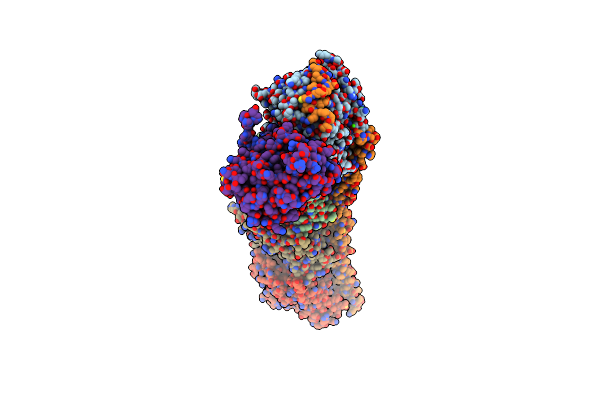 |
Organism: Mus musculus, Gallus gallus, Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-08-04 Classification: CELL CYCLE Ligands: GTP, MG, CA, CL, GDP, MES, GDF |