Search Count: 17
 |
Crystal Structure Of An Artificial Phytochrome Regulated Adenylate/Guanylate Cyclase In Its Dark Adapted Pr Form
Organism: Deinococcus radiodurans, Synechocystis sp. pcc 6803 substr. kazusa
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2018-04-18 Classification: LYASE Ligands: LBV, SO4, 12P |
 |
Organism: Beggiatoa sp. ps
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-04-05 Classification: LYASE Ligands: FMN, CL, CA |
 |
Organism: Beggiatoa sp. ps
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-04-05 Classification: LYASE Ligands: CL, FMN, MG |
 |
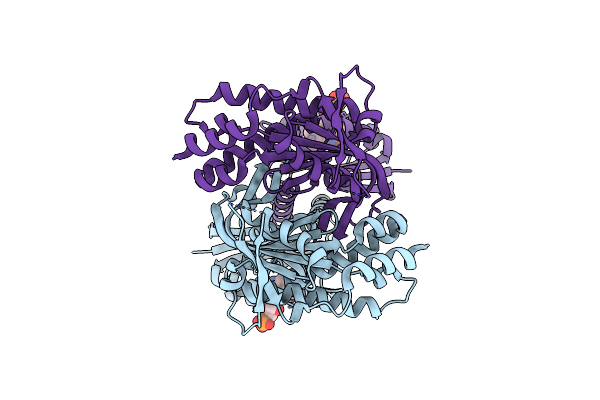
Organism: Beggiatoa sp. ps
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2017-04-05 Classification: LYASE Ligands: FMN |
 |
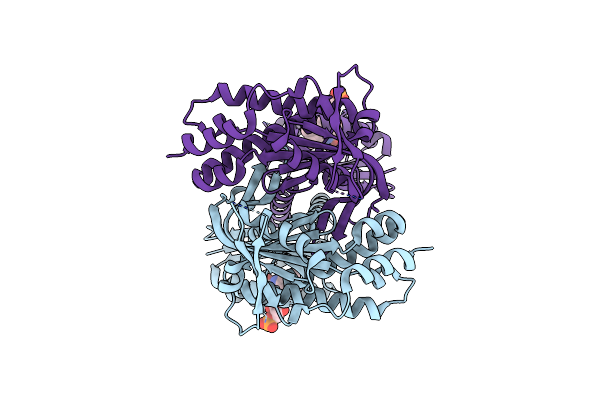
Organism: Beggiatoa sp. ps
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-04-05 Classification: LYASE Ligands: CL, FMN |
 |
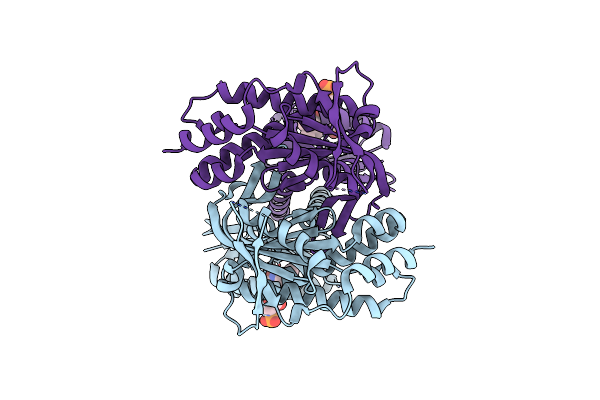
Organism: Beggiatoa sp. ps
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2017-04-05 Classification: LYASE Ligands: FMN, CL |
 |
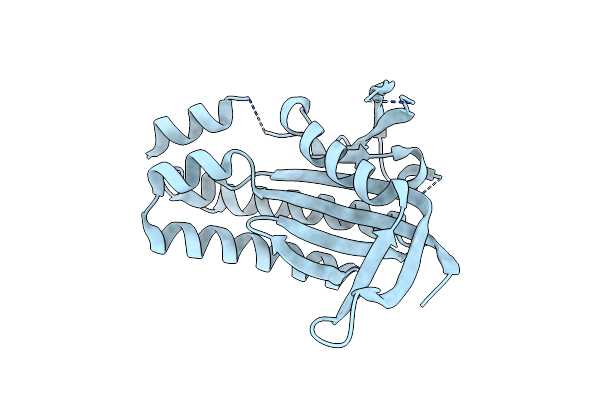
Organism: Beggiatoa sp. ps
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-04-05 Classification: LYASE Ligands: FMN |
 |
Organism: Beggiatoa sp. ps
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-04-05 Classification: LYASE Ligands: IOD |
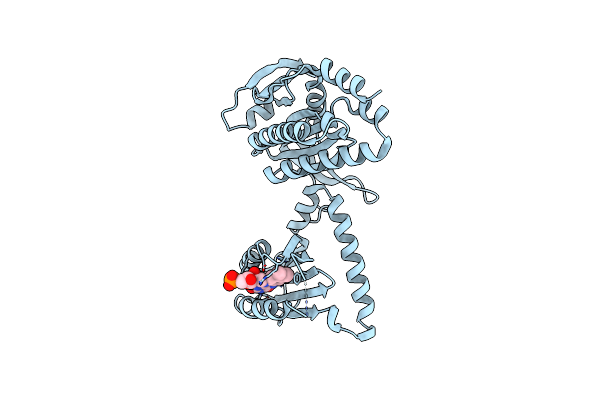 |
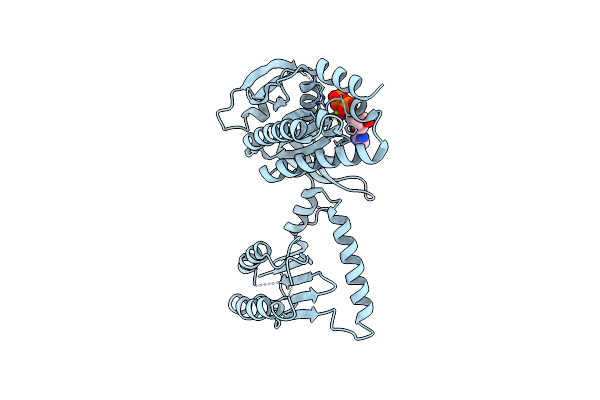
Organism: Beggiatoa sp. ps
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-04-05 Classification: LYASE Ligands: FMN |
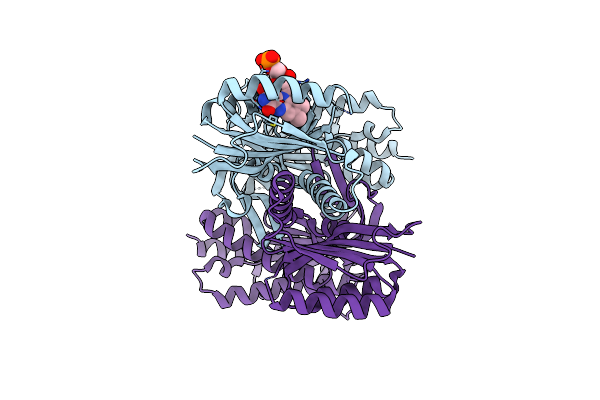 |
Organism: Beggiatoa sp. ps
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-04-05 Classification: LYASE Ligands: FMN |
 |
Organism: Beggiatoa sp. ps
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-04-05 Classification: LYASE Ligands: ATP, MG |
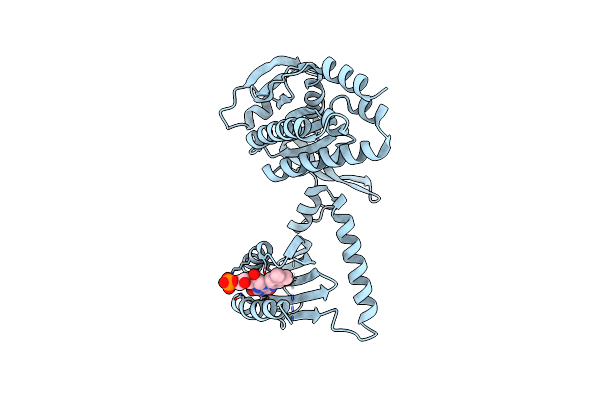 |
Organism: Beggiatoa sp. ps
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2017-04-05 Classification: LYASE Ligands: FMN |
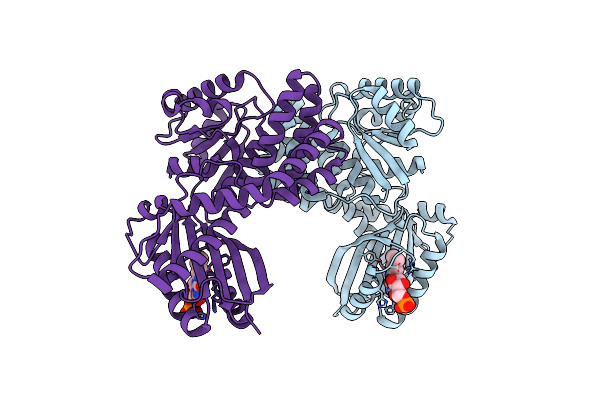 |
Dark-State Structure Of Appa C20S Without The Cys-Rich Region From Rb. Sphaeroides
Organism: Rhodobacter sphaeroides
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2013-06-05 Classification: FLAVOPROTEIN,SIGNALING PROTEIN Ligands: FMN, CL |
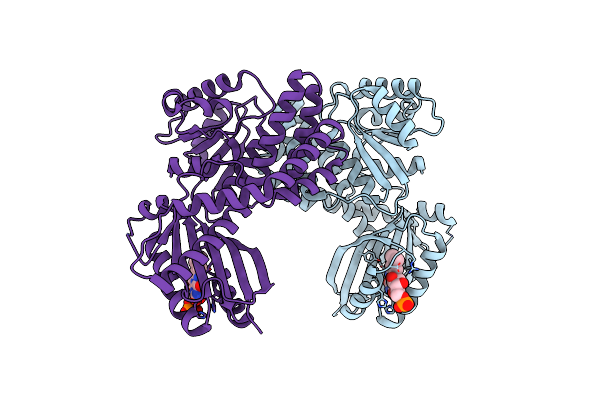 |
Dark-State Structure Of Appa Wild-Type Without The Cys-Rich Region From Rb. Sphaeroides
Organism: Rhodobacter sphaeroides
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2013-06-05 Classification: FLAVOPROTEIN,SIGNALING PROTEIN Ligands: FMN |
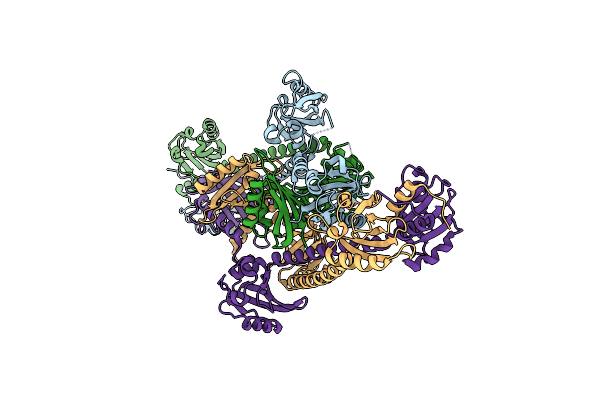 |
Organism: Rhodobacter sphaeroides
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2013-06-05 Classification: TRANSCRIPTION |
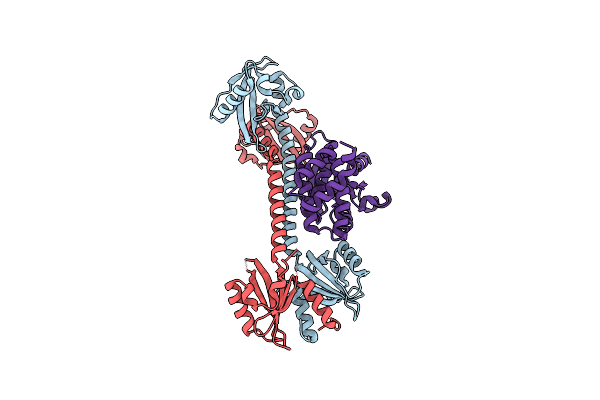 |
Organism: Rhodobacter sphaeroides
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2013-06-05 Classification: FLAVOPROTEIN/TRANSCRIPTION |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:3.06 Å Release Date: 2011-11-09 Classification: PROTEIN TRANSPORT Ligands: GDP, MG, AF3, SO4 |

