Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
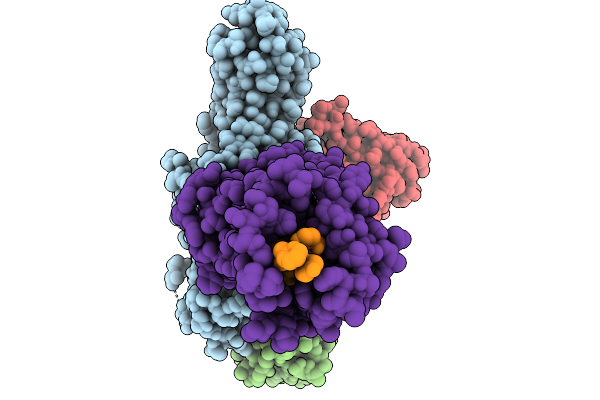 |
Cryo-Em Structure Of The G Protein-Coupled Receptor 1 (Gpr1) Bound To Chemerin And Beta-Arrestin 1 (Conformation 1)
Organism: Homo sapiens, Escherichia phage ecszw-2
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM |
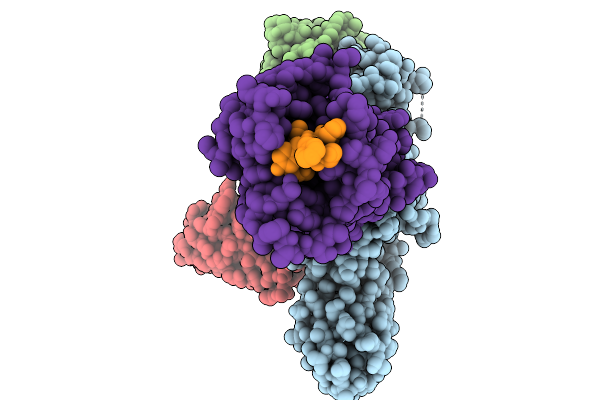 |
Cryo-Em Structure Of The G Protein-Coupled Receptor 1 (Gpr1) Bound To Chemerin And Beta-Arrestin 1 (Conformation 2)
Organism: Homo sapiens, Escherichia phage ecszw-2
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM |
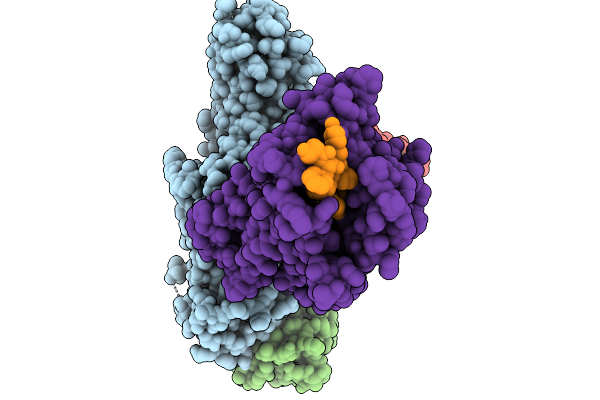 |
Cryo-Em Structure Of The G Protein-Coupled Receptor 1 (Gpr1) Bound To Chemerin And Beta-Arrestin 1 (Conformation 3)
Organism: Homo sapiens, Escherichia phage ecszw-2
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM |
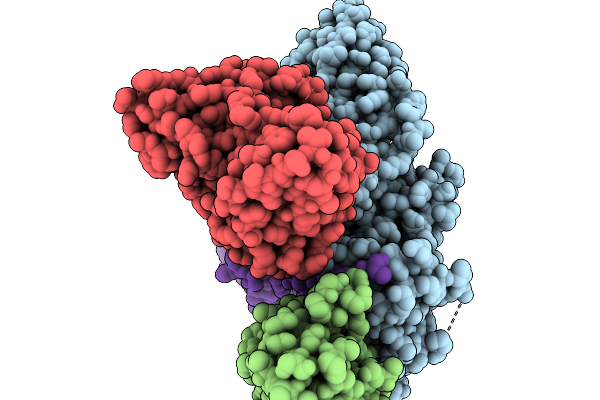 |
Cryo-Em Structure Of The G Protein-Coupled Receptor 1 (Gpr1) Bound To Chemerin And Beta-Arrestin 1 (Conformation 4)
Organism: Homo sapiens, Escherichia phage ecszw-2
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM |
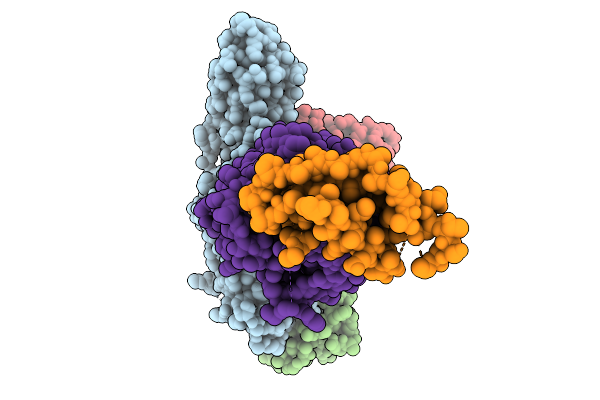 |
Composite Map Of The G Protein-Coupled Receptor 1 (Gpr1) Bound To Chemerin And Beta-Arrestin 2
Organism: Homo sapiens, Escherichia phage ecszw-2
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM Ligands: Y01 |
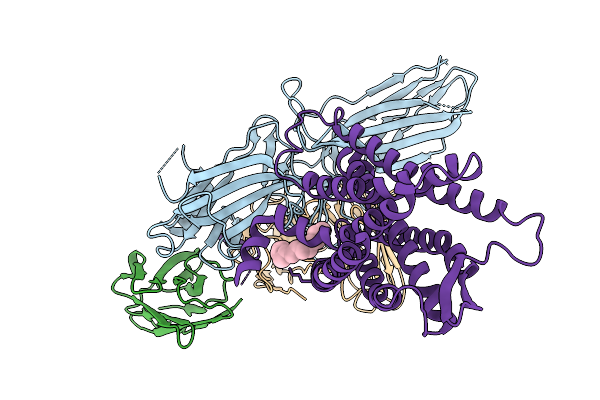 |
Cryo-Em Structure Of The G Protein-Coupled Receptor 1 (Gpr1) Bound To Beta-Arrestin 1 In Ligand-Free State
Organism: Homo sapiens, Escherichia phage ecszw-2
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM Ligands: PAM |
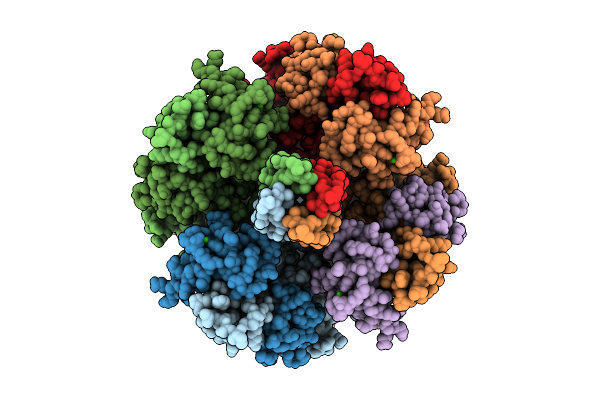 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: MEMBRANE PROTEIN Ligands: PT5, K, CA |
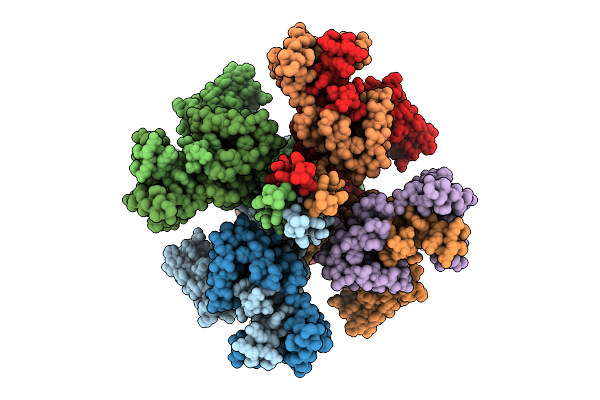 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: MEMBRANE PROTEIN Ligands: PIO, K |
 |
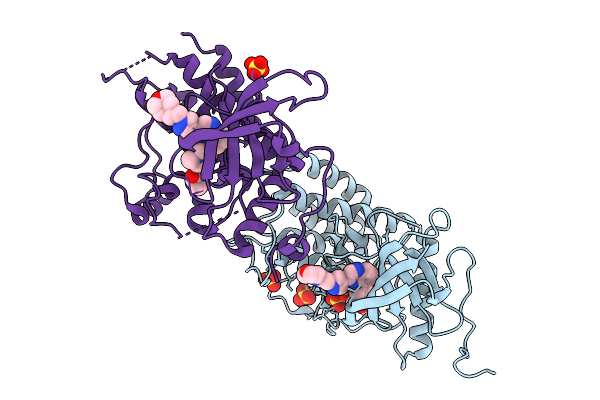
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: TRANSFERASE Ligands: A1EPE, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: TRANSFERASE Ligands: A1EPF, SO4 |
 |
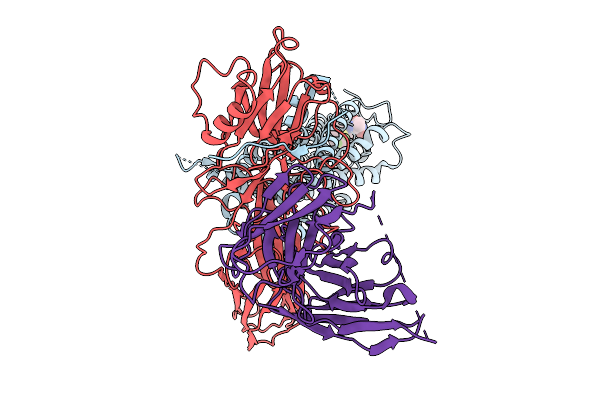
Mechanistic Insights Into The Versatile Stoichiometry And Biased Signaling Of The Apelin Receptor-Arrestin Complex
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: SIGNALING PROTEIN Ligands: A1L6R, CLR |
 |
Cryo-Em Structure Of Dimeric Apjr And Two Beta-Arrestins Complex With Small Molecules
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: SIGNALING PROTEIN Ligands: A1L6R, CLR |
 |
Cryo-Em Structure Of Dimeric Apjr And One Beta-Arrestin Complex With Small Molecules
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: SIGNALING PROTEIN Ligands: A1L6R |
 |
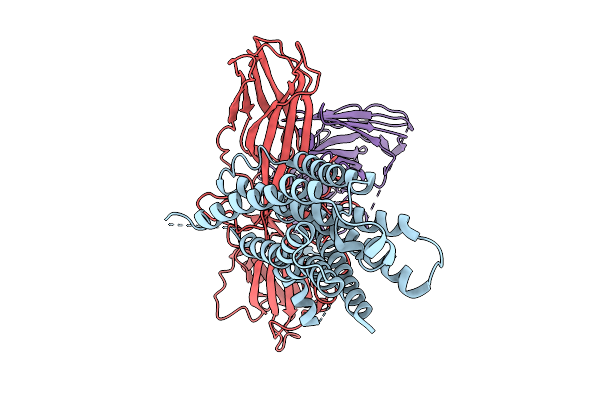
Cryo-Em Structure Of The Orphan Gpr52 Beta-Arrestin 1 Complex Bound To Ligand C17
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM Ligands: EN6 |
 |
Cryo-Em Structure Of The Orphan Gpr52 Bound To Beta-Arrestin 1 In Ligand-Free State
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM |
 |
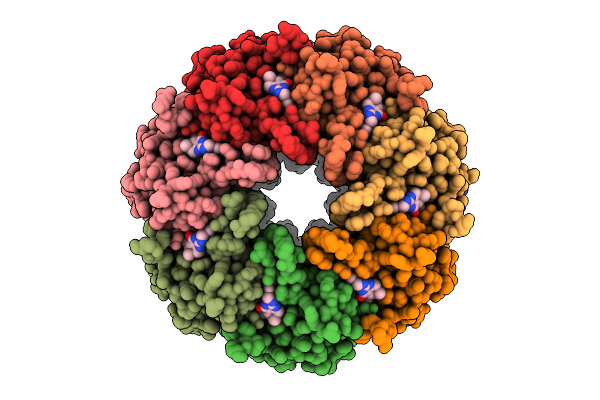
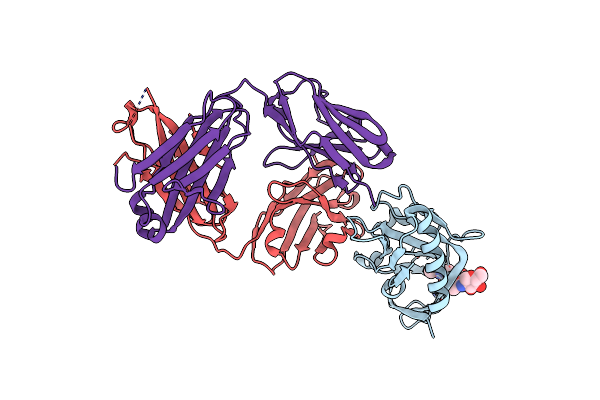
Crystal Structure Of Ljf-0085 Fab, A Non-Human Primate Antibody Targets Hiv-1 Envelope Protein
Organism: Macaca mulatta
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: ANTITUMOR PROTEIN Ligands: A1EG3 |
 |
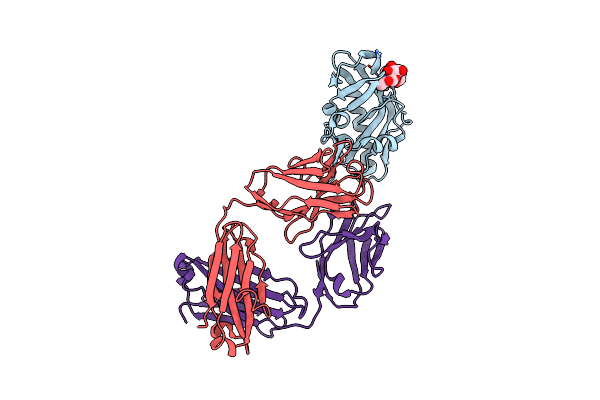
Crystal Structure Of Antibody Fab G001-0087 From Iavi G001 Human Trial In Complex With A Germline-Targeting Gp120 Engineered Outer Domain Eod-Gt8-Mingly
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: IMMUNE SYSTEM Ligands: NAG |
 |
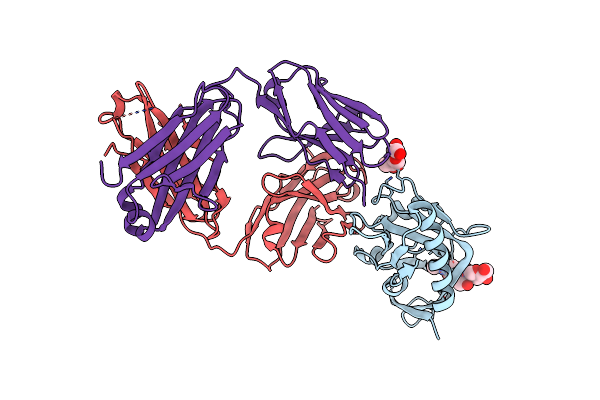
Crystal Structure Of Antibody Fab G001-58 From Iavi G001 Human Trial In Complex With A Germline-Targeting Gp120 Engineered Outer Domain Eod-Gt8-Mingly
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: IMMUNE SYSTEM Ligands: NAG |
 |
Crystal Structure Of Antibody Fab G001-59 From Iavi G001 Human Trial In Complex With A Germline-Targeting Gp120 Engineered Outer Domain Eod-Gt8-Mingly-N276
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: IMMUNE SYSTEM Ligands: NAG |