Search Count: 32
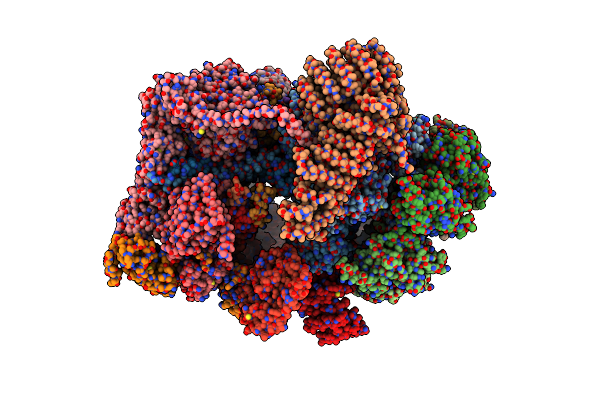 |
Cryo-Em Structure Of Human 26S Rp (Ed State) Bound To K11/K48-Branched Ubiquitin (Ub) Chain Composed Of Four Ub.
Organism: Homo sapiens, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: CYTOSOLIC PROTEIN Ligands: ATP, MG, ADP |
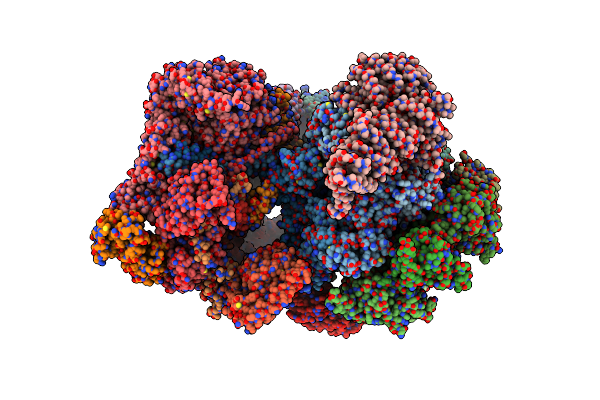 |
Cryo-Em Structure Of Human 26S Rp (Eb State) Bound To K11/K48-Branched Ubiquitin (Ub) Chain Composed Of Four Ub.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-12-25 Classification: CYTOSOLIC PROTEIN Ligands: ATP, MG, ADP |
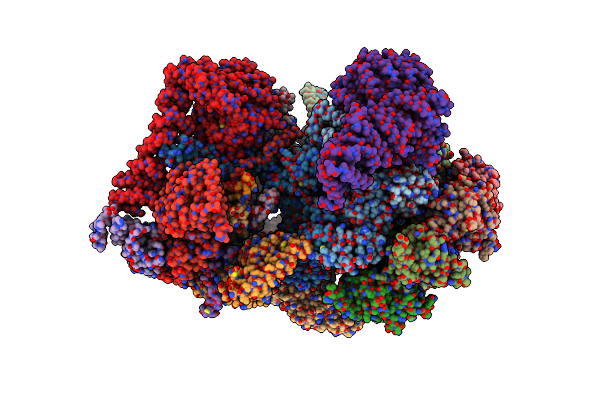 |
Cryo-Em Structure Of Human 26S Proteasomal Rp Subcomplex (Ea State) Without Any Bound Substrate.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: CYTOSOLIC PROTEIN Ligands: ATP, MG, ADP |
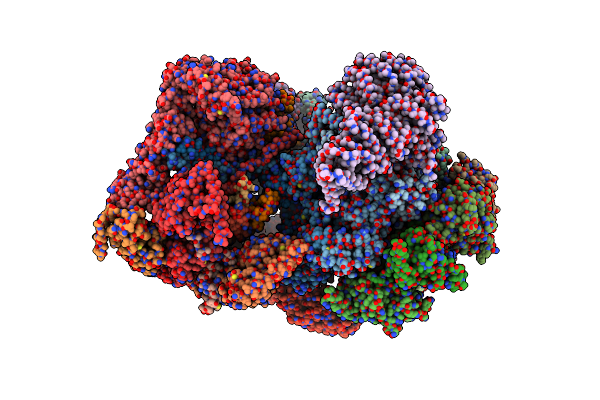 |
Cryo-Em Structure Of Human 26S Proteasomal Rp Subcomplex (Ea State) Bound To K11/K48-Branched Ubiquitin (Ub) Chain Composed Of Three Ub.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: CYTOSOLIC PROTEIN Ligands: ATP, MG, ADP |
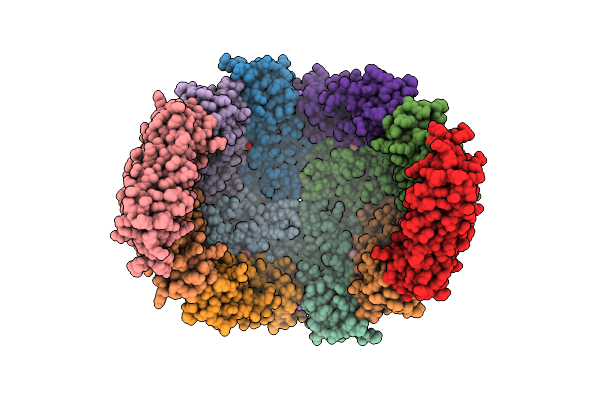 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2024-12-04 Classification: METAL BINDING PROTEIN |
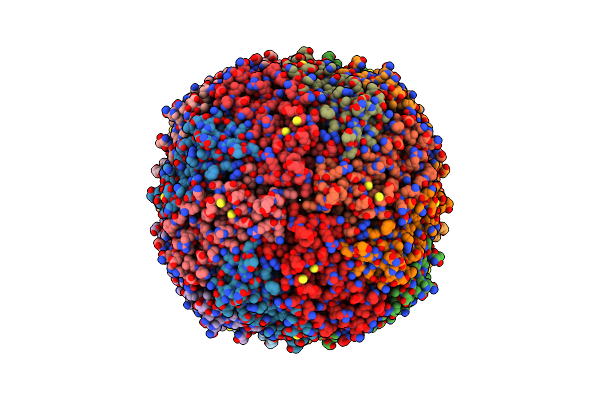 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-11-27 Classification: METAL BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-11-27 Classification: METAL BINDING PROTEIN Ligands: CU |
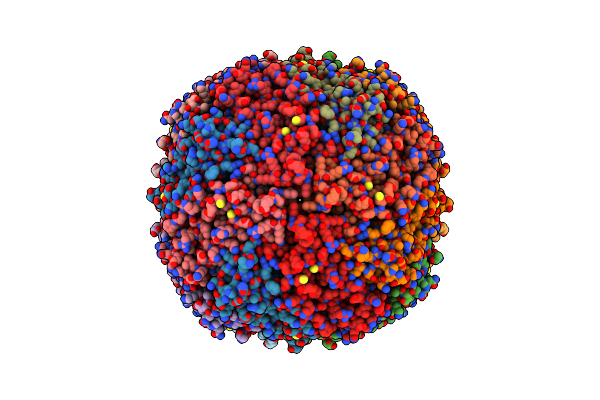 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-11-27 Classification: METAL BINDING PROTEIN Ligands: FE |
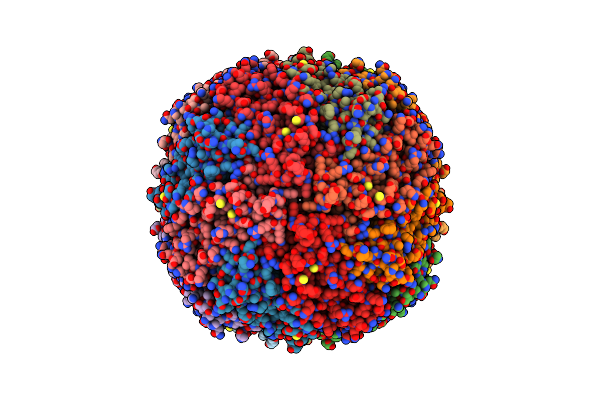 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-11-27 Classification: METAL BINDING PROTEIN Ligands: CU |
 |
Crystal Structure Of Transportin-1 In Complex With Bap1 Py-Nls (Residues 706-724)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.76 Å Release Date: 2022-04-20 Classification: PROTEIN TRANSPORT |
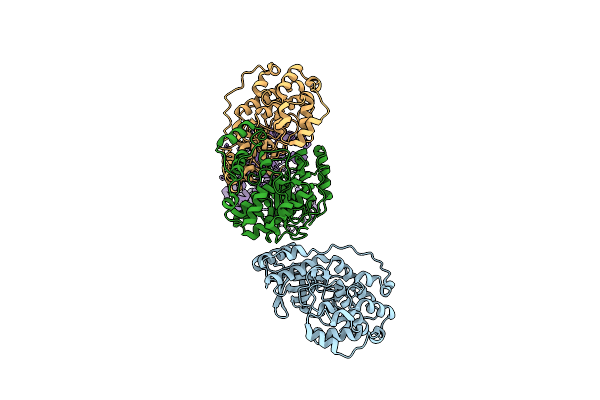 |
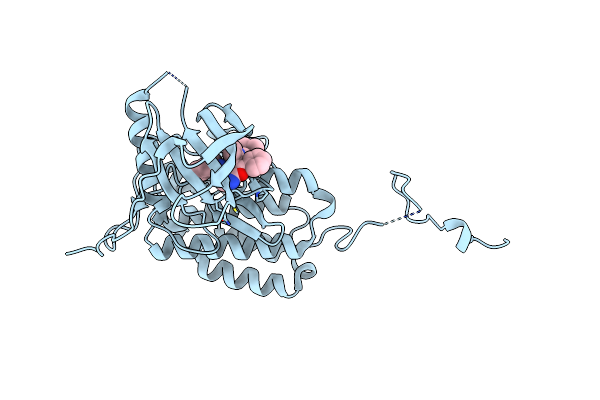
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2021-06-30 Classification: SIGNALING PROTEIN |
 |
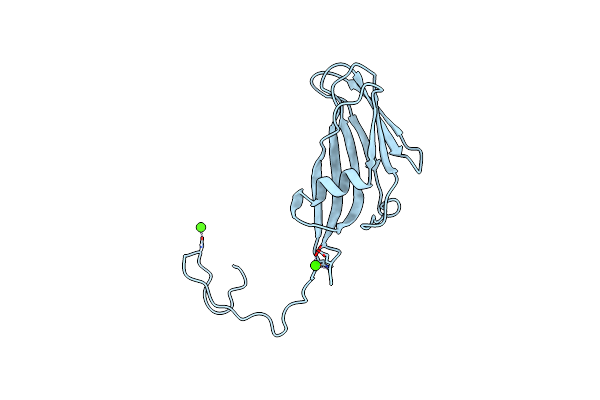
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.86 Å Release Date: 2019-12-04 Classification: TRANSFERASE/INHIBITOR Ligands: CKO |
 |
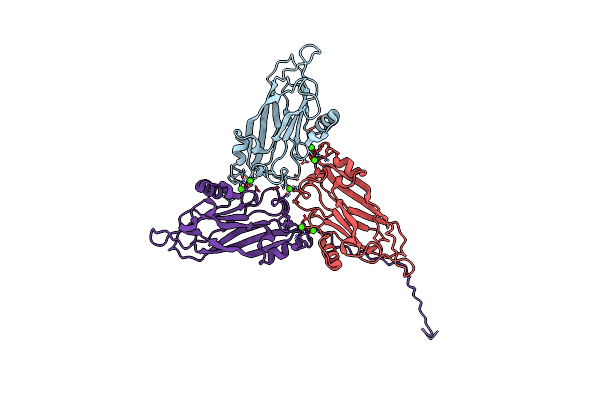
Cryo-Em Structure Of Giant Freshwater Prawn Macrobrachium Rosenbergii Extra Small Virus (Xsv) Vlp
Organism: Extra small virus
Method: ELECTRON MICROSCOPY Release Date: 2019-07-17 Classification: VIRUS Ligands: CA |
 |
Cryo-Em Structure Of Giant Freshwater Prawn Macrobrachium Rosenbergii Nodavirus (Mrnv) Semi-Empty Vlp
Organism: Macrobrachium rosenbergii nodavirus
Method: ELECTRON MICROSCOPY Release Date: 2019-05-01 Classification: VIRUS LIKE PARTICLE Ligands: CA |
 |
Cryo-Em Structure Of Giant Freshwater Prawn Macrobrachium Rosenbergii Nodavirus (Mrnv) Full Vlp
Organism: Macrobrachium rosenbergii nodavirus
Method: ELECTRON MICROSCOPY Release Date: 2019-05-01 Classification: VIRUS LIKE PARTICLE Ligands: CA |
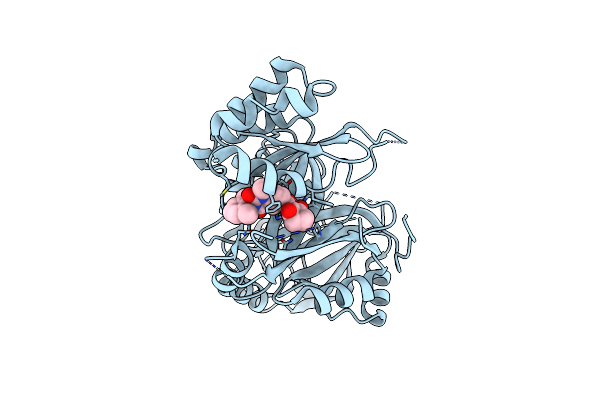 |
Crystal Structure Of Arabidopsis Thaliana Hppd Truncated Mutant Complexed With Benquitrione
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2019-01-16 Classification: OXIDOREDUCTASE Ligands: CO, 94L |
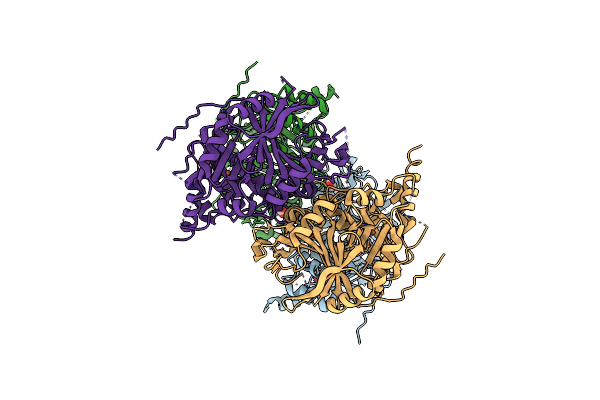 |
Crystal Structure Of Arabidopsis Thaliana 4-Hydroxyphenylpyruvate Dioxygenase (Athppd) Complexed With Its Substrate 4-Hydroxyphenylpyruvate Acid (Hppa)
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2018-09-19 Classification: OXIDOREDUCTASE Ligands: TYF, FE, ACT |
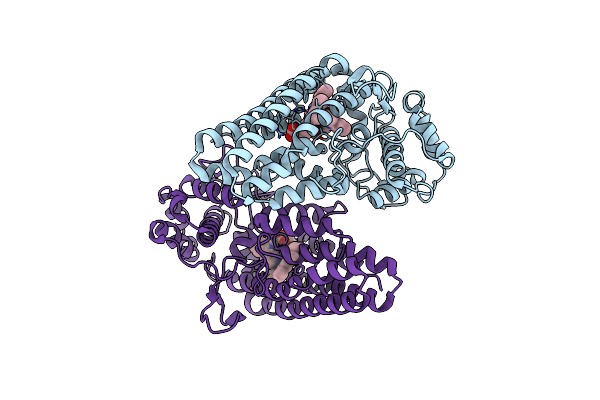 |
Crystal Structure Of The Indoleamine 2,3-Dioxygenagse 1 (Ido1) Complexed With Nlg919 Analogue
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2016-02-10 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: HEM, XNL |
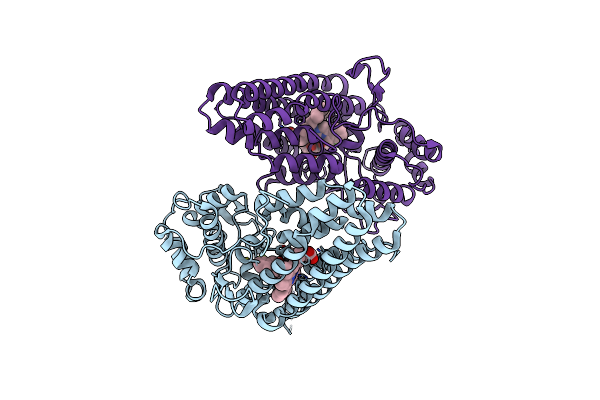 |
Crystal Structure Of The Indoleamine 2,3-Dioxygenagse 1 (Ido1) Complexed With Nlg919 Analogue
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2015-12-23 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: HEM, 5PJ |
 |
Crystal Structure Of The Indoleamine 2,3-Dioxygenagse 1 (Ido1) Complexed With Nlg919 Analogue
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2015-12-23 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: HEM, 5PK |

