Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
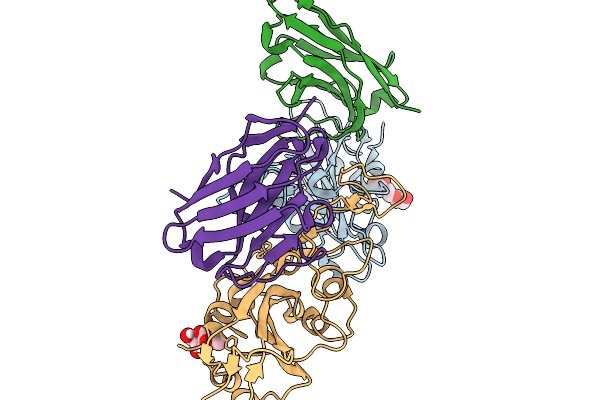 |
Crystal Structure Of Sars-Cov-2 Spike Receptor-Binding Domain (Delta) In Complex With Ph-Dependent Nanobody Mnb-11.
Organism: Severe acute respiratory syndrome coronavirus 2, Vicugna pacos
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: ANTIVIRAL PROTEIN Ligands: NAG |
 |
Organism: Monkeypox virus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: VIRAL PROTEIN |
 |
Cryo-Em Structure Of The Cytosolic Armh2-Efcab9-Catsperz Subcomplex Of The Mouse Catspermasome
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: CYTOSOLIC PROTEIN |
 |
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: SIGNALING PROTEIN Ligands: E2Q |
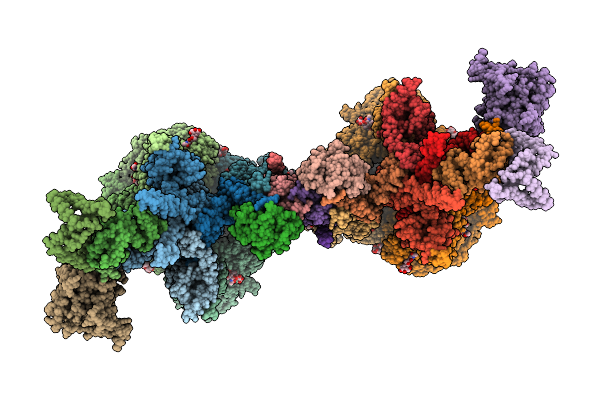 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: PROTEIN TRANSPORT Ligands: NAG, CLR |
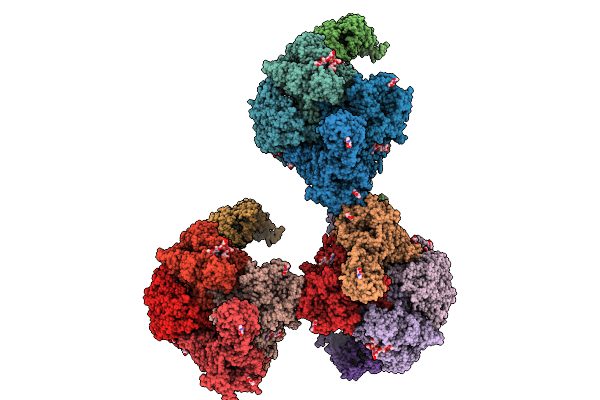 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: PROTEIN TRANSPORT Ligands: NAG, CLR |
 |
Organism: Monkeypox virus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: VIRAL PROTEIN |
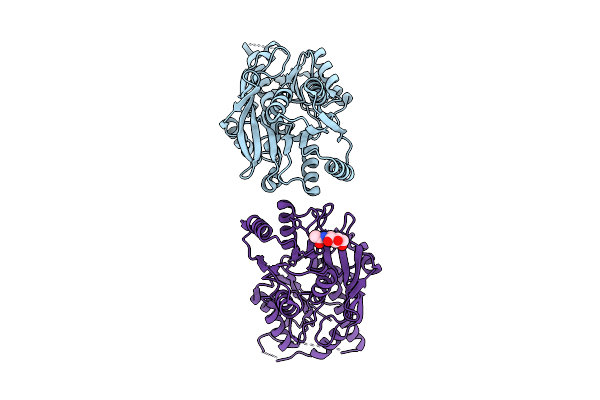 |
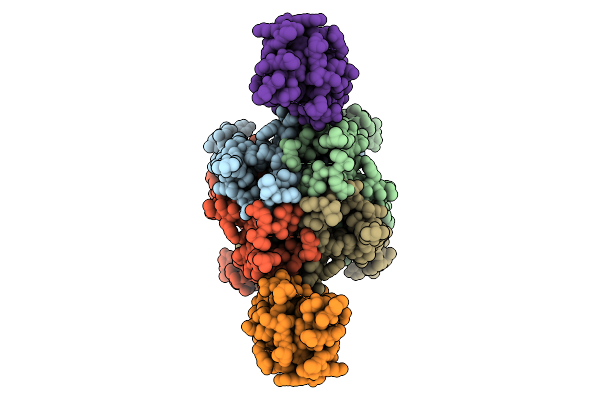
Cerebellar Glua2/4 Ntd Heterophilic Tetramer Interface (Focused Refinement)
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: SIGNALING PROTEIN Ligands: NAG |
 |
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: SIGNALING PROTEIN |
 |
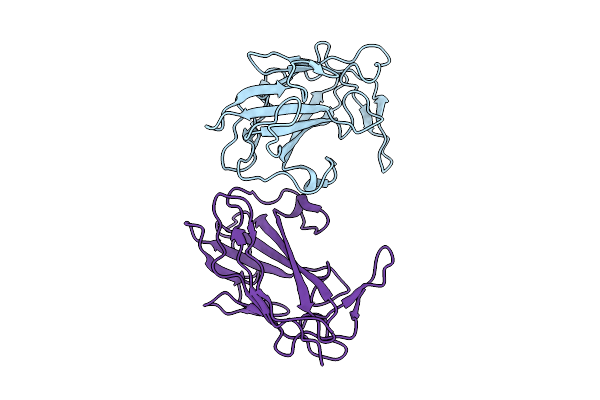
Structure Of Flagellar Hook Subunit Flge D-I Domain In Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: MOTOR PROTEIN |
 |
Structure Of Flagellar Hook Subunit Flge D-Ii Domain In Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: MOTOR PROTEIN |
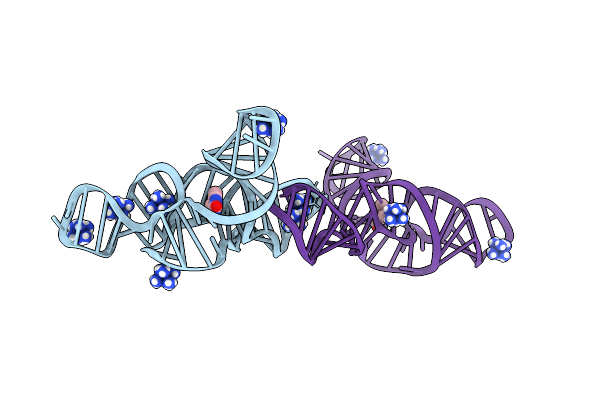 |
Structure Of The Bacillus Subtilis Yjdf Riboswitch Complexed With Lumichrome In The Presence Of Iridium Hexammine
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: RNA Ligands: LUM, IRI |
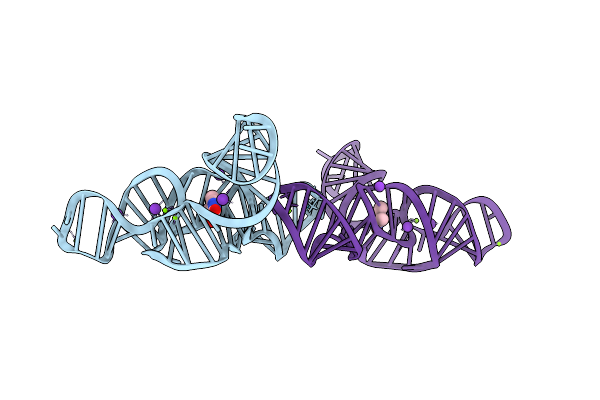 |
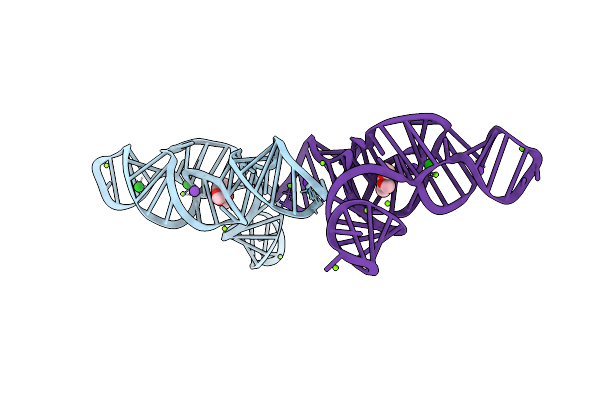
Structure Of The Bacillus Subtilis Yjdf Riboswitch Aptamer Domain In Complex With Lumichrome
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: RNA Ligands: MG, K, LUM |
 |
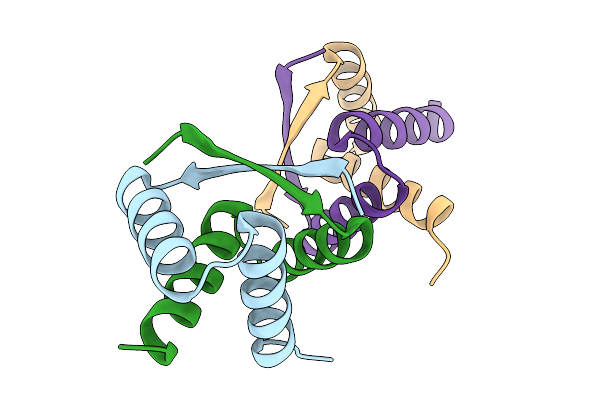
Structure Of The Bacillus Subtilis Yjdf Riboswitch Aptamer Domain In Complex With Chelerythrine
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: RNA Ligands: CTI, BA, K, MG |
 |
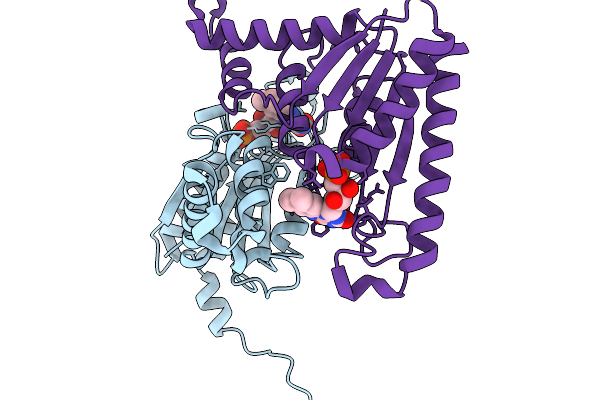
Organism: Streptomyces albus
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: DNA BINDING PROTEIN |
 |
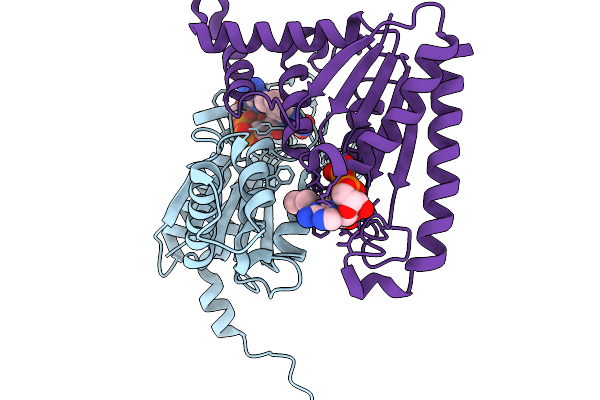
Organism: Streptomyces albus
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: FLAVOPROTEIN Ligands: FMN |
 |
Organism: Streptomyces albus
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: FLAVOPROTEIN Ligands: FAD |
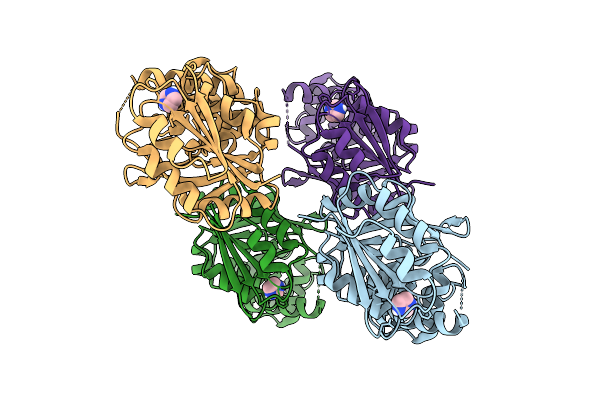 |
Organism: Rhizobium tropici ciat 899
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: TRANSFERASE Ligands: IMD, CL |
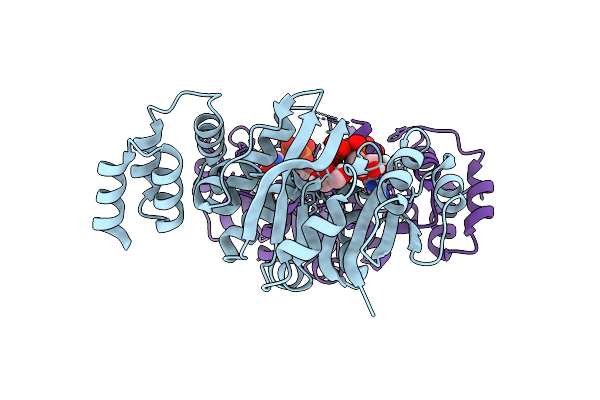 |
Organism: Rhizobium tropici ciat 899
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: TRANSFERASE Ligands: GDP, GOL, SO4 |
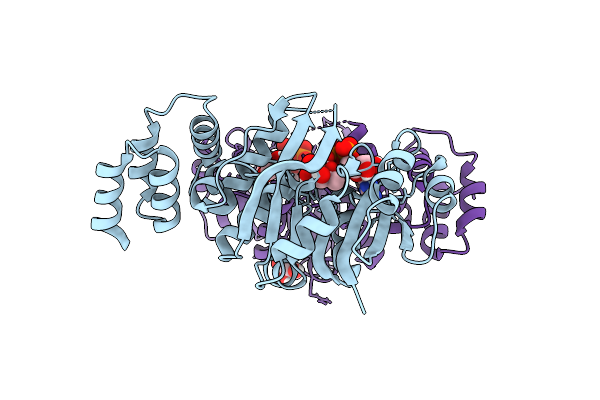 |
Organism: Rhizobium tropici ciat 899
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: TRANSFERASE Ligands: GFB, GOL |