Search Count: 14
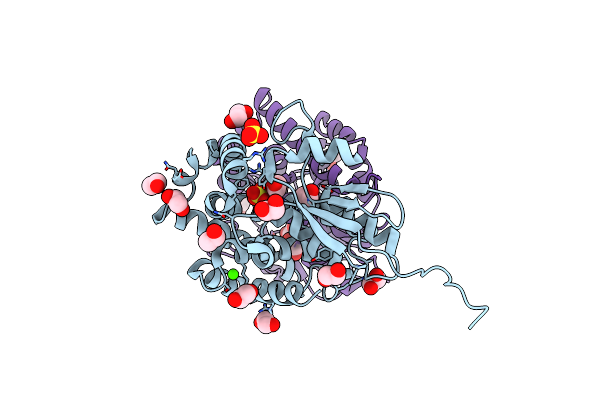 |
Crystal Structure Of An Apo Form Of The Glutathione S-Transferase, Csgst83044, Of Ceriporiopsis Subvermispora
Organism: Ceriporiopsis subvermispora (strain b)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2019-05-15 Classification: TRANSFERASE Ligands: EDO, CA, SO4 |
 |
Crystal Structure Of The Glutathione S-Transferase, Csgst83044, Of Ceriporiopsis Subvermispora In Complex With Glutathione
Organism: Ceriporiopsis subvermispora (strain b)
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2019-05-15 Classification: TRANSFERASE Ligands: GSH |
 |
Crystal Structure Of A Double Glycine Motif Protease From Ams/Pcat Transporter In Complex With The Leader Peptide
Organism: Lachnospiraceae bacterium c6a11, Prochlorococcus marinus str. mit 9313
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-02-06 Classification: TRANSPORT PROTEIN Ligands: 16P |
 |
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2016-08-10 Classification: TRANSFERASE Ligands: SF4, DTV |
 |
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2016-08-10 Classification: TRANSFERASE Ligands: SF4, DTV |
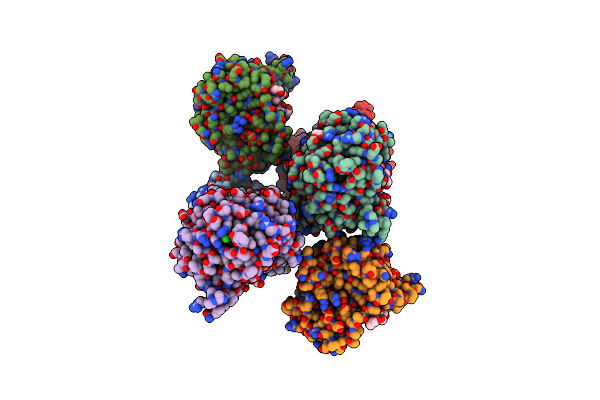 |
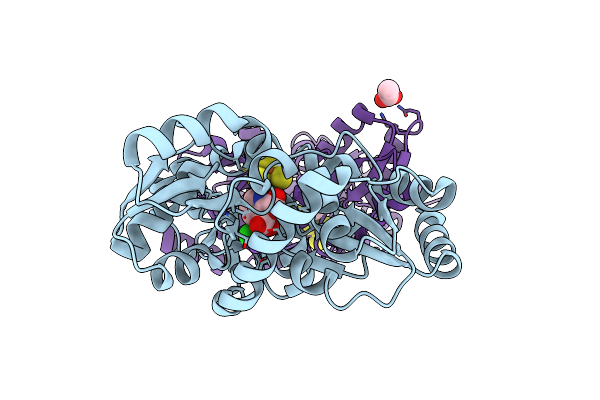
Crystal Structure Of M. Tuberculosis Lipoyl Synthase With 6-Thiooctanoyl Peptide Intermediate
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv), Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2016-08-10 Classification: TRANSFERASE Ligands: F3S, SF4, 5AD, MET, IMD, CL, IPA |
 |
Crystallographic Capture Of Quinolinate Synthase (Nada) From Pyrococcus Horikoshii In Its Substrates And Product-Bound States
Organism: Pyrococcus horikoshii (strain atcc 700860 / dsm 12428 / jcm 9974 / nbrc 100139 / ot-3)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-06-29 Classification: TRANSFERASE Ligands: SF4, NTM, CL, NA, ACT |
 |
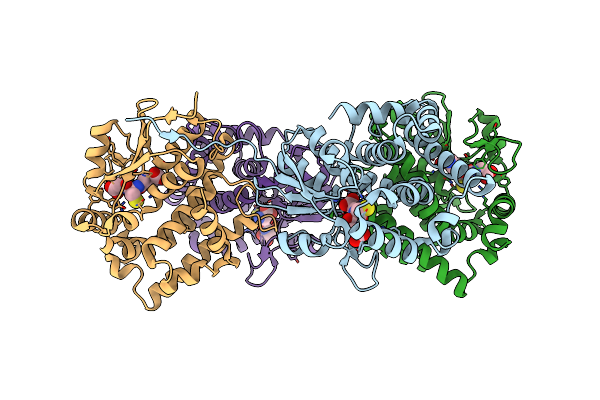
Low Resolution Crystal Structure Of Human Ribonucleotide Reductase Alpha6 Hexamer In Complex With Datp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:9.01 Å Release Date: 2016-01-20 Classification: OXIDOREDUCTASE |
 |
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2013-05-08 Classification: OXIDOREDUCTASE Ligands: SF4, SAM, CL |
 |
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2013-05-08 Classification: OXIDOREDUCTASE Ligands: SF4, SAM, GOL, CL |
 |
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2013-05-08 Classification: OXIDOREDUCTASE Ligands: SF4, SAM, GOL, CL |
 |
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2013-05-08 Classification: OXIDOREDUCTASE Ligands: SF4, SAM, GOL, CL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2011-05-11 Classification: OXIDOREDUCTASE Ligands: SF4, MRD |
 |
X-Ray Structure Of Rlmn From Escherichia Coli In Complex With S-Adenosylmethionine
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2011-05-11 Classification: OXIDOREDUCTASE Ligands: SF4, SAM |

