Search Count: 40
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2025-02-12 Classification: IMMUNE SYSTEM Ligands: PEU, IPA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-21 Classification: RNA BINDING PROTEIN Ligands: ZN |
 |
Organism: Drosophila melanogaster
Method: ELECTRON MICROSCOPY Release Date: 2024-08-21 Classification: RNA BINDING PROTEIN Ligands: ZN |
 |
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-08-23 Classification: TRANSFERASE Ligands: ADN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2023-08-02 Classification: IMMUNE SYSTEM Ligands: P15 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2023-03-01 Classification: IMMUNE SYSTEM Ligands: P15 |
 |
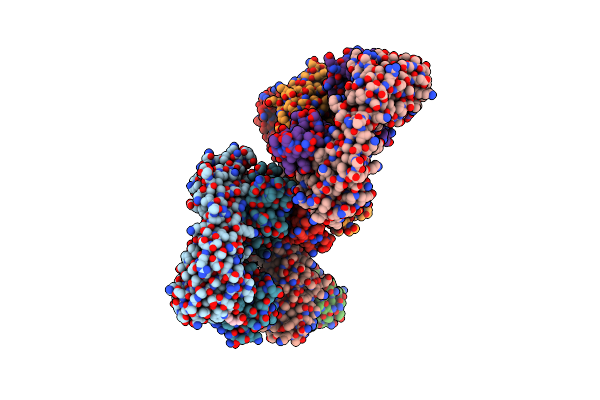
Cryo-Em Structure Of Drosophila Integrator Cleavage Module (Ints4-Ints9-Ints11) In Complex With Ip6
Organism: Drosophila melanogaster
Method: ELECTRON MICROSCOPY Release Date: 2022-10-12 Classification: NUCLEASE Ligands: ZN, IHP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2022-08-10 Classification: IMMUNE SYSTEM Ligands: P15 |
 |
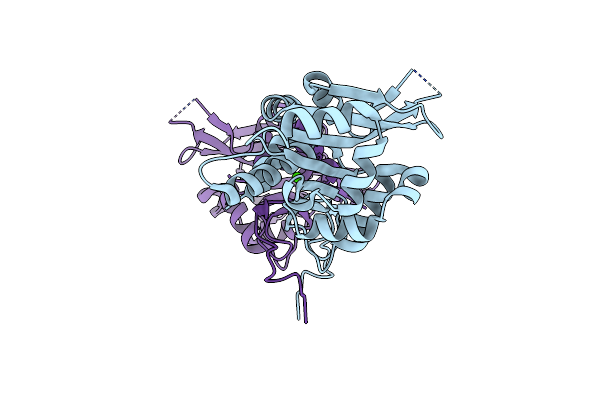
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2021-08-18 Classification: VIRAL PROTEIN |
 |
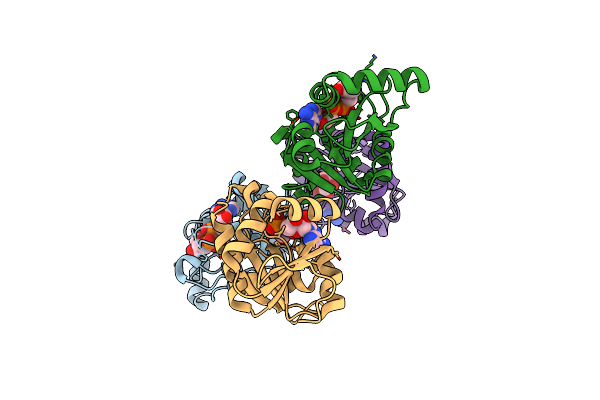
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2020-12-09 Classification: CHAPERONE Ligands: CA |
 |
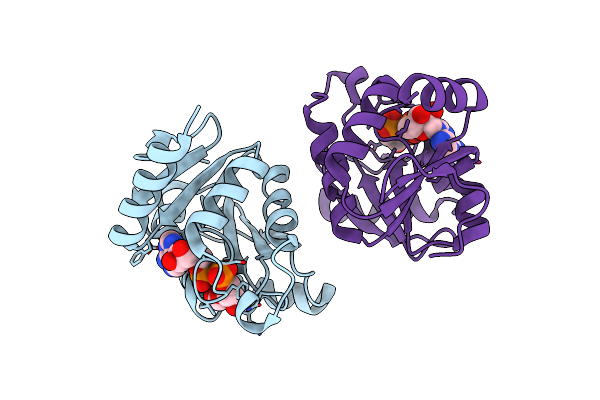
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:3.83 Å Release Date: 2020-11-11 Classification: HYDROLASE Ligands: APR |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2020-11-11 Classification: VIRAL PROTEIN Ligands: APR |
 |
Crystal Structure Of Poly(Adp-Ribose) Glycohydrolase (Parg) From Deinococcus Radiodurans In Apo Form
Organism: Deinococcus radiodurans (strain atcc 13939 / dsm 20539 / jcm 16871 / lmg 4051 / nbrc 15346 / ncimb 9279 / r1 / vkm b-1422)
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2019-02-27 Classification: HYDROLASE Ligands: SO4 |
 |
Crystal Structure Of Poly(Adp-Ribose) Glycohydrolase (Parg) From Deinococcus Radiodurans In Complex With Adp-Ribose (P21)
Organism: Deinococcus radiodurans (strain atcc 13939 / dsm 20539 / jcm 16871 / lmg 4051 / nbrc 15346 / ncimb 9279 / r1 / vkm b-1422)
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2019-02-27 Classification: HYDROLASE Ligands: AR6 |
 |
Crystal Structure Of Poly(Adp-Ribose) Glycohydrolase (Parg) From Deinococcus Radiodurans In Complex With Adp-Ribose (P32)
Organism: Deinococcus radiodurans r1
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2019-02-27 Classification: HYDROLASE Ligands: AR6, PO4 |
 |
Crystal Structure Of Poly(Adp-Ribose) Glycohydrolase (Parg) From Deinococcus Radiodurans In Complex With Adp-Ribose (P212121)
Organism: Deinococcus radiodurans (strain atcc 13939 / dsm 20539 / jcm 16871 / lmg 4051 / nbrc 15346 / ncimb 9279 / r1 / vkm b-1422)
Method: X-RAY DIFFRACTION Resolution:2.73 Å Release Date: 2019-02-27 Classification: HYDROLASE Ligands: AR6, PO4 |
 |
Crystal Structure Of Poly(Adp-Ribose) Glycohydrolase (Parg) From Deinococcus Radiodurans In Complex With Adp-Ribose (P3221)
Organism: Deinococcus radiodurans r1
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2019-02-27 Classification: HYDROLASE Ligands: AR6 |
 |
Crystal Structure Of Poly(Adp-Ribose) Glycohydrolase (Parg) T267K Mutant From Deinococcus Radiodurans In Complex With Adp-Ribose
Organism: Deinococcus radiodurans (strain atcc 13939 / dsm 20539 / jcm 16871 / lmg 4051 / nbrc 15346 / ncimb 9279 / r1 / vkm b-1422)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-02-27 Classification: HYDROLASE Ligands: AR6 |
 |
Crystal Structure Of Poly(Adp-Ribose) Glycohydrolase (Parg) T267R Mutant From Deinococcus Radiodurans In Complex With Adp-Ribose
Organism: Deinococcus radiodurans (strain atcc 13939 / dsm 20539 / jcm 16871 / lmg 4051 / nbrc 15346 / ncimb 9279 / r1 / vkm b-1422)
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2019-02-27 Classification: HYDROLASE Ligands: AR6 |
 |
Organism: Trichomonas vaginalis
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2018-07-18 Classification: ISOMERASE |

