Search Count: 429
 |
Organism: Caenorhabditis elegans
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: MEMBRANE PROTEIN |
 |
Organism: Caenorhabditis elegans
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: MEMBRANE PROTEIN |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: SIGNALING PROTEIN Ligands: CA, CAC, GOL, CL |
 |
Organism: Mus musculus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: SIGNALING PROTEIN Ligands: CA |
 |
Organism: Mus musculus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: SIGNALING PROTEIN Ligands: CA |
 |
Organism: Acinetobacter baylyi adp1
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: PROTEIN BINDING Ligands: ACT, CD |
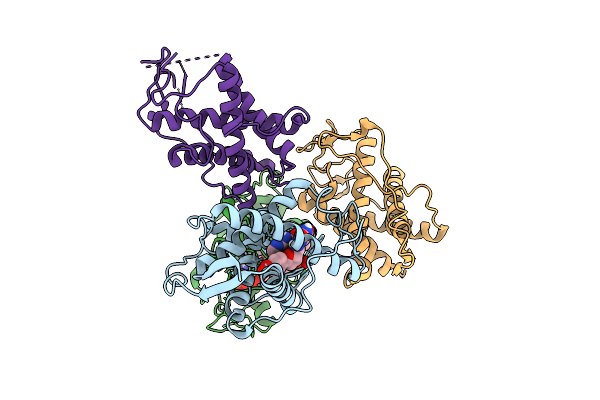 |
The Crystal Structure Of Human Ryr3 Repeat12 Domain In Complex With Dantrolene And Amp-Pcp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: U1C, ACP |
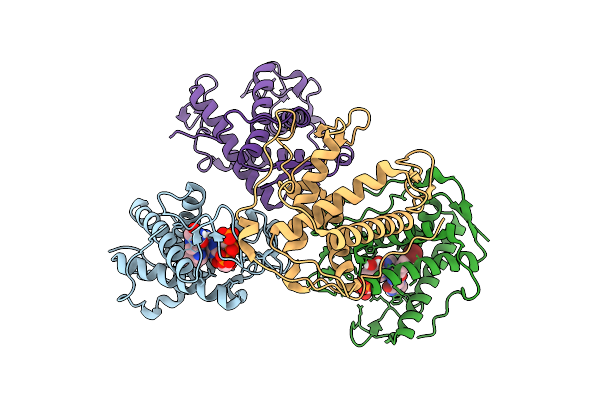 |
The Crystal Structure Of Human Ryr3 Repeat12 Domain In Complex With Azumolene And Amp-Pcp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: ACP, A1L7B |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: GOL |
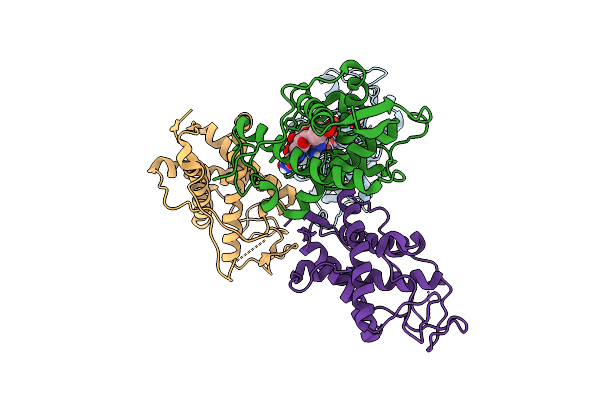 |
The Crystal Structure Of Human Ryr3 Repeat12 Domain In Complex With Dantrolene And Adp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: ADP, U1C |
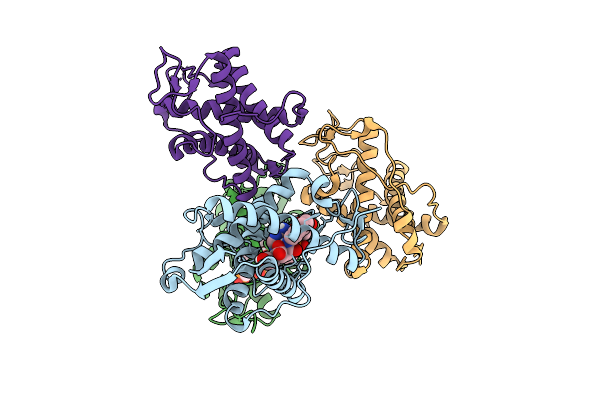 |
The Crystal Structure Of Human Ryr3 Repeat12 Domain Complex With Ri-2 And Amp-Pcp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: ACP, A1L7G |
 |
Organism: Homo sapiens, Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: RIBOSOME Ligands: ZN, MG |
 |
Organism: Micromonospora echinaurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: OXIDOREDUCTASE Ligands: NAP |
 |
Organism: Micromonospora echinaurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: OXIDOREDUCTASE |
 |
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: MOTOR PROTEIN Ligands: ADP, VO4, MG, DMS, EDO, A1IGE, CL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: ENDOCYTOSIS Ligands: CA, NAG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: ENDOCYTOSIS Ligands: NAG, CA, HEM, OXY |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: ENDOCYTOSIS Ligands: CA, HEM, OXY |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: HYDROLASE Ligands: A1IIH, EDO |

