Search Count: 18
 |
Cryoem Structure Of Delta Opioid Receptor Bound To G Proteins And A Partial Agonist
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Resolution:2.80 Å Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: A1AWC |
 |
Cryoem Structure Of Delta Opioid Receptor Bound To G Proteins And A Full Bitopic Agonist
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Resolution:2.62 Å Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: A1AWD |
 |
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: SIGNALING PROTEIN/IMMUNE SYSTEM Ligands: A1AQ2 |
 |
Organism: Lama glama, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2024-07-17 Classification: MEMBRANE PROTEIN Ligands: A1APV, A1APU |
 |
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2022-12-07 Classification: MEMBRANE PROTEIN Ligands: KZR |
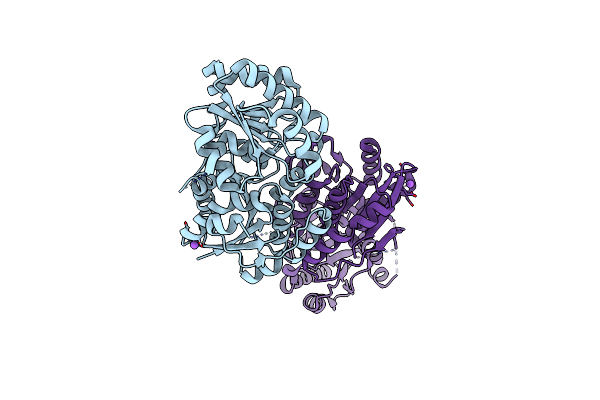 |
Organism: Streptomyces kasugaensis
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2022-11-16 Classification: ISOMERASE |
 |
The Crystal Structure Of Non-Hydrolyzing Udpglcnac 2-Epimerase In Complex With Udp-Glucose
Organism: Streptomyces kasugaensis
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2022-11-16 Classification: ISOMERASE Ligands: UPG, NA |
 |
The Crystal Structure Of Non-Hydrolyzing Udpglcnac 2-Epimerase In Complex With Udp
Organism: Streptomyces kasugaensis
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2022-11-16 Classification: ISOMERASE Ligands: UDP, NA |
 |
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2022-05-04 Classification: MEMBRANE PROTEIN Ligands: L0X |
 |
Structural And Mechanistic Studies Of A Novel Non-Heme Iron Epimerase/Lyase And Its Utilization In Chemoselective Synthesis.
Organism: Streptomyces albus
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2022-03-16 Classification: ISOMERASE Ligands: FE, GOA, 5RN, JAX |
 |
Structural And Mechanistic Studies Of A Novel Non-Heme Iron Epimerase/Lyase And Its Utilization In Chemoselective Synthesis.
Organism: Streptomyces albus
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2022-03-16 Classification: ISOMERASE Ligands: PO3, FE |
 |
Structural And Mechanistic Studies Of A Novel Non-Heme Iron Epimerase/Lyase And Its Utilization In Chemoselective Synthesis.
Organism: Streptomyces albus
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2022-03-16 Classification: ISOMERASE Ligands: FE, 3IO |
 |
Structural And Mechanistic Studies Of A Novel Non-Heme Iron Epimerase/Lyase And Its Utilization In Chemoselective Synthesis.
Organism: Streptomyces albus
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2022-03-16 Classification: ISOMERASE Ligands: FE, JCX |
 |
Structural And Mechanistic Studies Of A Novel Non-Heme Iron Epimerase/Lyase And Its Utilization In Chemoselective Synthesis.
Organism: Streptomyces albus
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2022-03-16 Classification: ISOMERASE Ligands: PO3, FE, JD3 |
 |
Structural And Mechanistic Studies Of A Novel Non-Heme Iron Epimerase/Lyase And Its Utilization In Chemoselective Synthesis.
Organism: Streptomyces albus
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2022-03-16 Classification: ISOMERASE Ligands: I3A, FE, GOA |
 |
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.47 Å Release Date: 2020-05-20 Classification: PROTEIN BINDING Ligands: NAG, EPE |
 |
Organism: Gluconobacter oxydans
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2015-04-15 Classification: OXIDOREDUCTASE |
 |
Organism: Gluconobacter oxydans
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2015-04-15 Classification: OXIDOREDUCTASE |

