Search Count: 34
 |
Organism: Mus musculus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2021-01-13 Classification: HYDROLASE/DNA |
 |
Organism: Mus musculus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2021-01-13 Classification: HYDROLASE/DNA |
 |
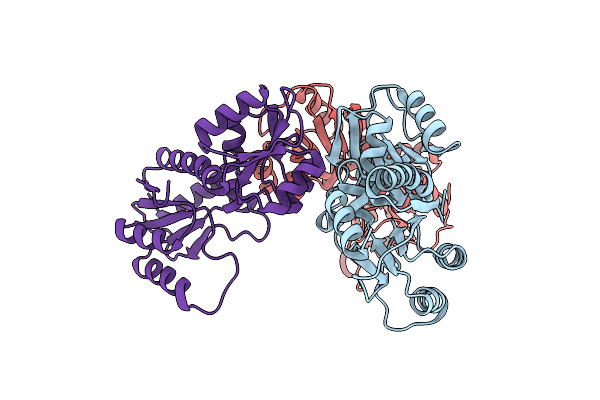
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-11-07 Classification: DNA BINDING PROTEIN Ligands: PO4 |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-11-07 Classification: DNA BINDING PROTEIN Ligands: MG, DCM, CA |
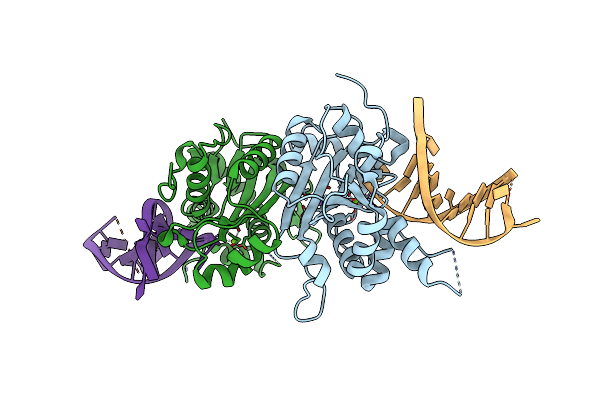 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-11-07 Classification: DNA BINDING PROTEIN Ligands: MG, NA |
 |
Structure Of Trex2 In Complex With A Duplex Dna With 2 Nucleotide 3'-Overhang
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2018-11-07 Classification: DNA BINDING PROTEIN Ligands: MG |
 |
Crystal Structure Of The Conserved Hypothetical Protein Mj0927 From Methanocaldococcus Jannaschii (In C2221 Form)
Organism: Methanocaldococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:2.47 Å Release Date: 2014-01-29 Classification: UNKNOWN FUNCTION |
 |
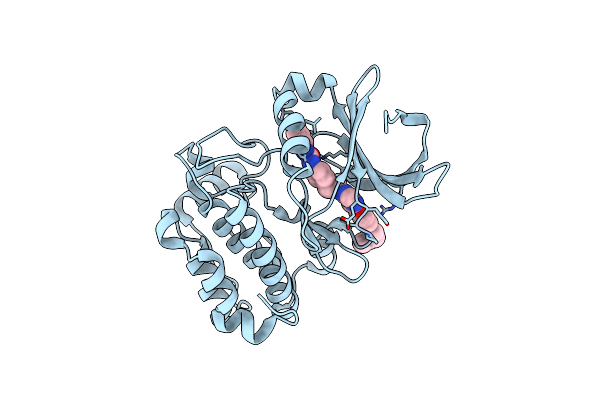
Crystal Structure Of The Conserved Hypothetical Protein Mj0927 From Methanocaldococcus Jannaschii (In P21 Form)
Organism: Methanocaldococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2014-01-29 Classification: UNKNOWN FUNCTION |
 |
Novel Aurora Kinase Inhibitors Reveal Mechanisms Of Hurp In Nucleation Of Centrosomal And Kinetochore Microtubules
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2013-06-05 Classification: TRANSFERASE Ligands: WPH |
 |
Novel Aurora Kinase Inhibitors Reveal Mechanisms Of Hurp In Nucleation Of Centrosomal And Kinetochore Microtubules
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2013-06-05 Classification: TRANSFERASE Ligands: YPH |
 |
Novel Aurora Kinase Inhibitors Reveal Mechanisms Of Hurp In Nucleation Of Centrosomal And Kinetochore Microtubules
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-06-05 Classification: TRANSFERASE Ligands: VX6 |
 |
Organism: Vaccinia virus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2013-03-06 Classification: VIRAL PROTEIN Ligands: PEG |
 |
Crystal Structure Of Beta-Glucosidase From Termite Neotermes Koshunensis In Complex With Gluconolactone
Organism: Neotermes koshunensis
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2012-07-04 Classification: HYDROLASE Ligands: LGC, EPE, GOL, NA, CL |
 |
Crystal Structure Of Beta-Glucosidase From Termite Neotermes Koshunensis In Complex With 1-Deoxynojirimycin
Organism: Neotermes koshunensis
Method: X-RAY DIFFRACTION Resolution:0.99 Å Release Date: 2012-07-04 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: NOJ, EPE, GOL, NA, CL |
 |
Crystal Structure Of Beta-Glucosidase From Termite Neotermes Koshunensis In Complex With Glycerol
Organism: Neotermes koshunensis
Method: X-RAY DIFFRACTION Resolution:1.03 Å Release Date: 2012-07-04 Classification: HYDROLASE Ligands: GOL, NA, CL |
 |
Crystal Structure Of Beta-Glucosidase From Termite Neotermes Koshunensis In Complex With Bis-Tris
Organism: Neotermes koshunensis
Method: X-RAY DIFFRACTION Resolution:0.97 Å Release Date: 2012-07-04 Classification: HYDROLASE Ligands: BTB, GOL, NA |
 |
Crystal Structure Of Beta-Glucosidase From Termite Neotermes Koshunensis In Complex With Glucose
Organism: Neotermes koshunensis
Method: X-RAY DIFFRACTION Resolution:1.03 Å Release Date: 2012-07-04 Classification: HYDROLASE Ligands: BGC, GOL, NA, CL |
 |
Crystal Structure Of Beta-Glucosidase From Termite Neotermes Koshunensis In Complex With Cellobiose
Organism: Neotermes koshunensis
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2012-07-04 Classification: HYDROLASE Ligands: EDO, NA, CL |
 |
Crystal Structure Of Beta-Glucosidase From Termite Neotermes Koshunensis In Complex With Salicin
Organism: Neotermes koshunensis
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2012-07-04 Classification: HYDROLASE Ligands: SA0, SA9, EDO, NA, CL |
 |
Crystal Structure Of Beta-Glucosidase From Termite Neotermes Koshunensis In Complex With A New Glucopyranosidic Product
Organism: Neotermes koshunensis
Method: X-RAY DIFFRACTION Resolution:1.03 Å Release Date: 2012-07-04 Classification: HYDROLASE Ligands: GBH, GOL, NA, CL |

