Search Count: 21
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: CHAPERONE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2022-05-11 Classification: HYDROLASE |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2018-10-17 Classification: NUCLEAR PROTEIN Ligands: ADP, BEF, MG, ZN |
 |
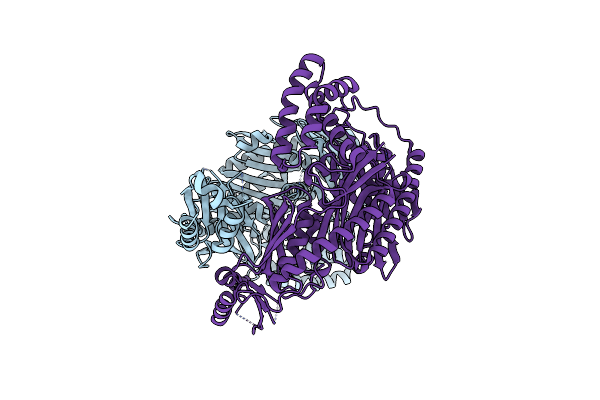
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2018-10-17 Classification: NUCLEAR PROTEIN Ligands: ADP, BEF, MG, ZN |
 |
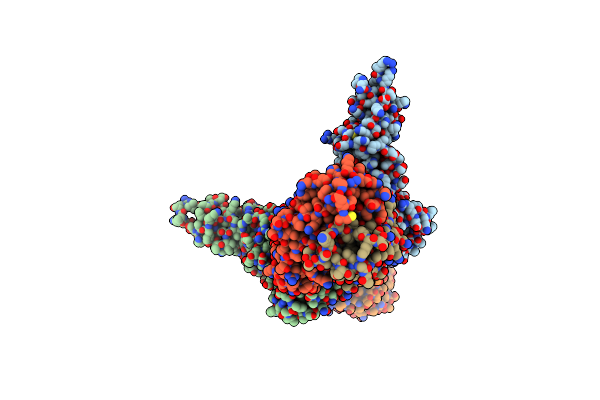
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2018-08-22 Classification: TRANSFERASE |
 |
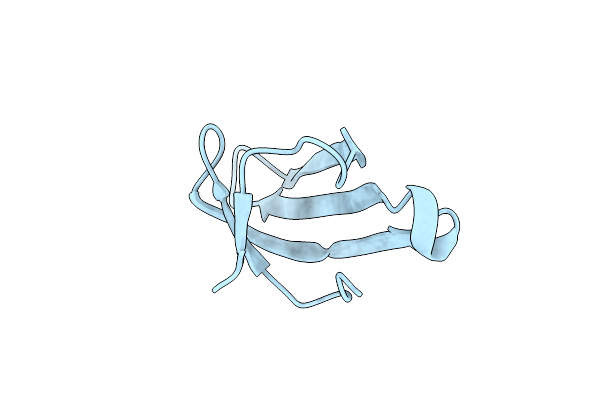
Crystal Structure Of Fab S40 In Complex With Influenza Hemagglutinin, Ha1 Subunit.
Organism: Influenza a virus (a/california/07/2009(h1n1)), Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2017-11-15 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
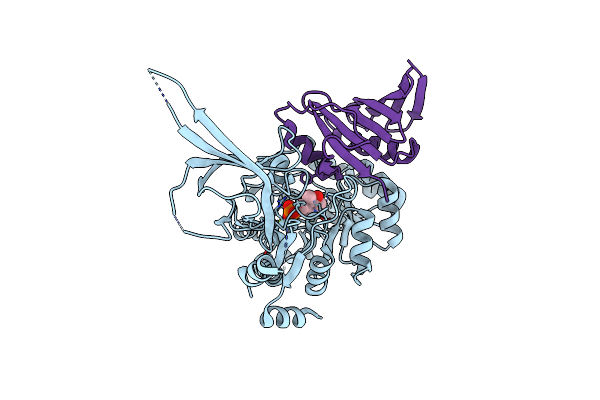
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2016-09-28 Classification: HYDROLASE |
 |
Structure Of Human Ornithine Decarboxylase In Complex With A C-Terminal Fragment Of Antizyme
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.63 Å Release Date: 2015-09-02 Classification: LYASE/LYASE INHIBITOR Ligands: PLP, MG |
 |
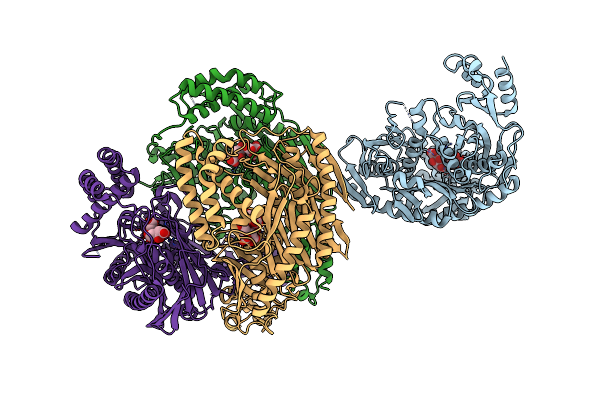
Structure Of Human Antizyme Inhibitor In Complex With A C-Terminal Fragment Of Antizyme
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:5.81 Å Release Date: 2015-09-02 Classification: PROTEIN BINDING |
 |
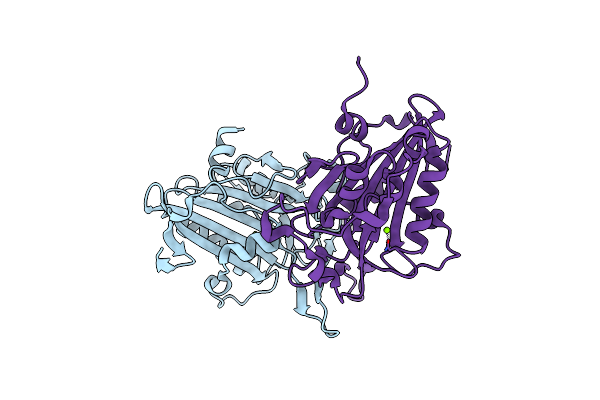
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2012-02-01 Classification: TRANSFERASE Ligands: CIT |
 |
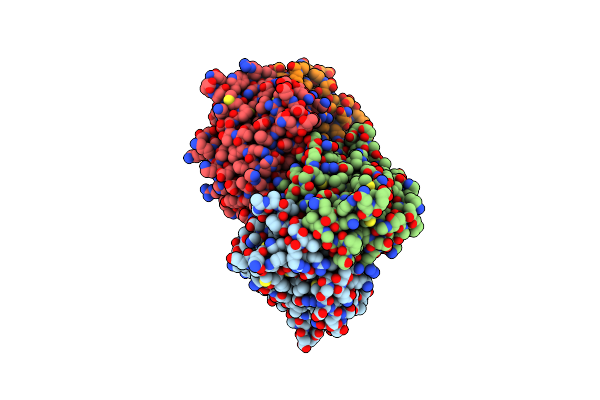
Crystal Structure Of Ced-3 Protease Suppressor-6 (Cps-6) From Caenorhabditis Elegans
Organism: Caenorhabditis elegans
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-01-11 Classification: HYDROLASE Ligands: MG |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2011-02-16 Classification: HYDROLASE Ligands: CO |
 |
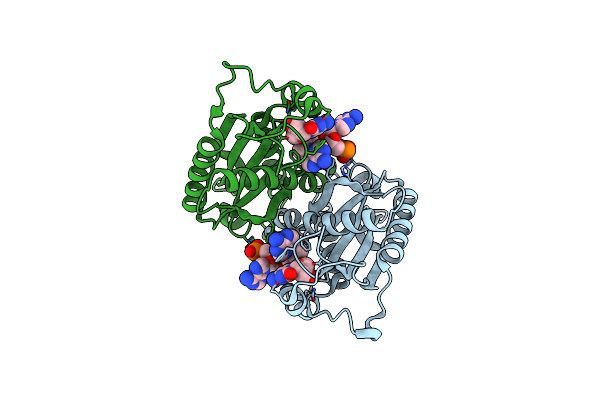
Crystal Structure Of Rnase T In Complex With A Non-Preferred Ssdna (Gc) With One Mg In The Active Site
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2011-02-16 Classification: HYDROLASE/DNA Ligands: MG, CO |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2011-02-16 Classification: HYDROLASE/DNA |
 |
Crystal Structure Of Rnase T In Complex With A Preferred Ssdna (Tagg) With Two Mg In The Active Site
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2011-02-16 Classification: HYDROLASE/DNA Ligands: MG |
 |
Organism: Escherichia coli, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2011-02-16 Classification: HYDROLASE/DNA |
 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2003-09-30 Classification: HYDROLASE Ligands: ZN |
 |
Structure Of The D55N Mutant Of Phospholipase C From Bacillus Cereus In Complex With (3S)-3,4,Di-N-Hexanoyloxybutyl-1-Phosphocholine
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2003-09-30 Classification: HYDROLASE Ligands: ZN, 3PC |
 |
Structure Of The D55N Mutant Of Phospholipase C From Bacillus Cereus In Complex With 1,2-Di-N-Pentanoyl-Sn-Glycero-3-Dithiophosphocholine
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2003-09-30 Classification: HYDROLASE Ligands: ZN, PC5 |
 |
Organism: Rattus norvegicus
Method: SOLUTION NMR Release Date: 2000-08-09 Classification: TRANSPORT PROTEIN Ligands: RTL |

