Search Count: 801
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2026-01-28 Classification: PROTEIN BINDING Ligands: NAD |
 |
Cryo-Em Structure Of The Vaccinia Virus Entry/Fusion Complex (Efc) Lacking The F9 Subunit
Organism: Orthopoxvirus vaccinia
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: VIRAL PROTEIN |
 |
Cryo-Em Structure Of The Vaccinia Virus Entry/Fusion Complex (Efc) Including The F9 Subunit
Organism: Orthopoxvirus vaccinia
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: VIRAL PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-12-10 Classification: HYDROLASE Ligands: ZN, PPS, EDO, ACY |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-12-10 Classification: HYDROLASE Ligands: MG, GDP |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-12-10 Classification: HYDROLASE Ligands: MG, GTP |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2025-12-10 Classification: HYDROLASE Ligands: MG, GTP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-12-10 Classification: HYDROLASE Ligands: MG, GNP |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSPORT PROTEIN Ligands: LMT |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSPORT PROTEIN |
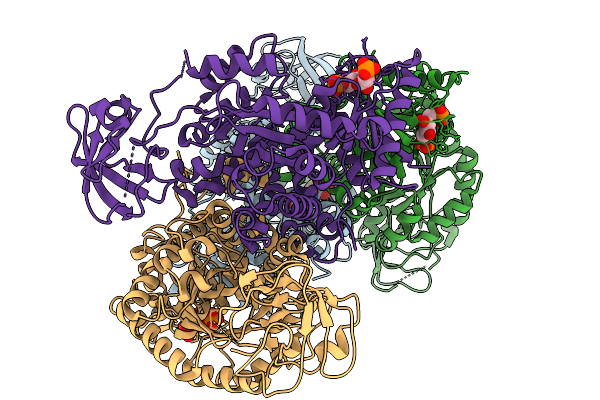 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2025-11-19 Classification: TRANSFERASE Ligands: GSH, FBP |
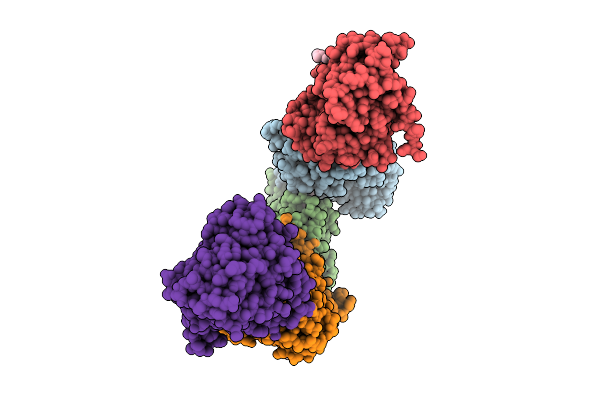 |
Crystal Structure Of Egfr Exon20 Insertion Mutant In Complex With Enozertinib (Oric-114)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2025-11-12 Classification: SIGNALING PROTEIN Ligands: A1L8T |
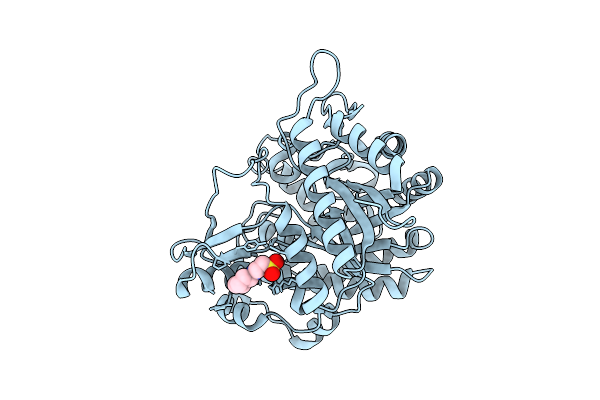 |
Neutron Structure Of Gh1 Beta-Glucosidase Td2F2 Ligand-Free Form At Room Temperature
Organism: Metagenome
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.19 Å, 1.8 Å Release Date: 2025-10-29 Classification: HYDROLASE Ligands: NHE, DOD |
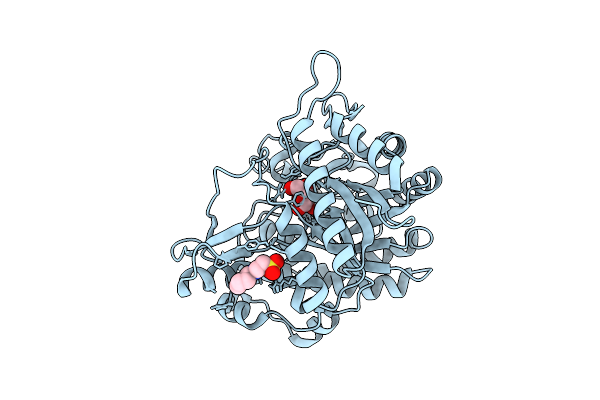 |
Neutron Structure Of Gh1 Beta-Glucosidase Td2F2 Glucose Complex At Room Temperature
Organism: Metagenome
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.20 Å, 1.70 Å Release Date: 2025-10-29 Classification: HYDROLASE Ligands: BGC, NHE, DOD |
 |
Neutron Structure Of Gh1 Beta-Glucosidase Td2F2 2F-Glc Complex At Room Temperature
Organism: Metagenome
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.32 Å, 1.70 Å Release Date: 2025-10-29 Classification: HYDROLASE Ligands: G2F, NHE, DOD |
 |
X-Ray Structure Of Gh1 Beta-Glucosidase Td2F2 2F-Glc Complex At Room Temperature
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:1.32 Å Release Date: 2025-10-29 Classification: HYDROLASE Ligands: G2F, NHE |
 |
X-Ray Structure Of Gh1 Beta-Glucosidase Td2F2 2F-Glc Complex At Cryogenic Temperature
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:1.11 Å Release Date: 2025-10-29 Classification: HYDROLASE Ligands: G2F, NA, GOL, NHE |
 |
Organism: Streptomyces cacaoi
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2025-10-22 Classification: OXIDOREDUCTASE Ligands: ZN, GOL |
 |
X-Ray Crystal Structure Of Streptomyces Cacaoi Polf In Complex With Iron And L-Isoleucine
Organism: Streptomyces cacaoi
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2025-10-15 Classification: OXIDOREDUCTASE Ligands: GOL, FE2, ZN, ILE |
 |
Structure Of A De Novo Designed Interleukin-21 Mimetic Complex With Il-21R And Il-2Rg
Organism: Synthetic construct, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2025-09-24 Classification: DE NOVO PROTEIN/IMMUNE SYSTEM Ligands: NAG, EDO |

