Search Count: 126
All
Selected
 |
Glycosyltransferase C From The Limosilactobacillus Reuteri Accessory Secretion System. Apo Form.
Organism: Limosilactobacillus reuteri
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-04-23 Classification: TRANSFERASE Ligands: CA, GOL |
 |
Glycosyltransferase C From The Limosilactobacillus Reuteri Accessory Secretion System. Complex With Udp-Glcnac.
Organism: Limosilactobacillus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-04-23 Classification: TRANSFERASE Ligands: UDP, CIT, GOL, EPE, CL, UD1 |
 |
Glycosyltransferase C From The Limosilactobacillus Reuteri Accessory Secretion System. Complex With Udp.
Organism: Limosilactobacillus reuteri
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2025-04-23 Classification: TRANSFERASE Ligands: UDP |
 |
Crystal Structure L-Lactate Dehydrogenase From Lactobacillus Reuteri In Its Apoform
Organism: Limosilactobacillus reuteri
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2025-04-02 Classification: OXIDOREDUCTASE Ligands: EDO |
 |
Structure Of Two Consecutively Arrayed Rib Domains (Rib8-9) From Surface Adhesin Of Limosilactobacillus Reuteri
Organism: Limosilactobacillus reuteri
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-03-05 Classification: STRUCTURAL PROTEIN Ligands: TRS |
 |
Organism: Limosilactobacillus reuteri
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2025-01-22 Classification: HYDROLASE Ligands: CA, GOL |
 |
Crystal Structure Of Inulosucrase From Lactobacillus Reuteri 121 In Complex With Fructose
Organism: Limosilactobacillus reuteri
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2025-01-22 Classification: HYDROLASE Ligands: FRU, CA |
 |
Crystal Structure Of Inulosucrase From Lactobacillus Reuteri 121 Mutant R544W
Organism: Limosilactobacillus reuteri
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2025-01-22 Classification: HYDROLASE Ligands: CA, EDO, GOL, 1PE, PEG |
 |
Crystal Structure Of Cysteine Synthase A From Limosilactobacillus Reuteri Lr1 In Its Apo Form
Organism: Limosilactobacillus reuteri
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-11-13 Classification: TRANSFERASE Ligands: PGE, EDO, PEG, SO4 |
 |
Crystal Structure Of Glucose 1-Dehydrogenase Mutant1 From Limosilactobacillus Fermentum
Organism: Limosilactobacillus fermentum
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2024-10-30 Classification: HYDROLASE |
 |
Crystal Structure Of Glucose 1-Dehydrogenase Mutant2 From Limosilactobacillus Fermentum
Organism: Limosilactobacillus fermentum
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2024-10-30 Classification: HYDROLASE |
 |
Crystal Structure Of Glucose 1-Dehydrogenase From Limosilactobacillus Fermentum
Organism: Limosilactobacillus fermentum
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-10-30 Classification: HYDROLASE Ligands: MG |
 |
Crystal Structure Of A Short-Chain Dehydrogenase From Lactobacillus Fermentum With Nadph
Organism: Limosilactobacillus fermentum
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2024-10-30 Classification: HYDROLASE Ligands: NAP |
 |
Structure Of Rib Domain From Surface Adhesin Of Limosilactobacillus Reuteri
Organism: Limosilactobacillus reuteri
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2024-09-11 Classification: STRUCTURAL PROTEIN Ligands: CU, NA, EDO |
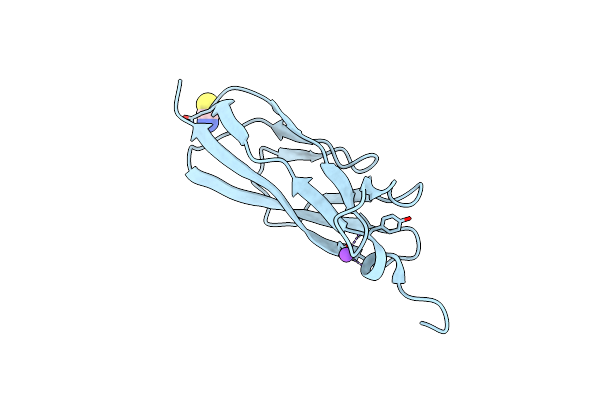 |
Organism: Limosilactobacillus reuteri
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2024-05-08 Classification: STRUCTURAL PROTEIN |
 |
Organism: Limosilactobacillus reuteri
Method: X-RAY DIFFRACTION Resolution:2.66 Å Release Date: 2024-01-03 Classification: TRANSFERASE |
 |
Organism: Limosilactobacillus reuteri
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2024-01-03 Classification: TRANSFERASE |
 |
Organism: Limosilactobacillus reuteri
Method: X-RAY DIFFRACTION Resolution:2.76 Å Release Date: 2023-12-27 Classification: TRANSFERASE |
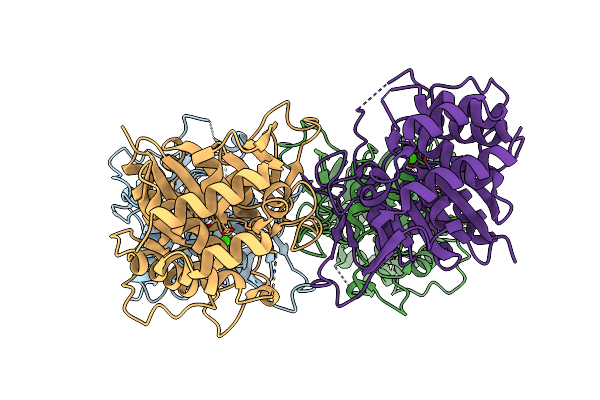 |
Crystal Structure Of The Ribonucleoside Hydrolase C From Lactobacillus Reuteri
Organism: Limosilactobacillus reuteri
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-12-20 Classification: HYDROLASE Ligands: CA |
 |
Conversion Of Indole-3-Acetic Acid Into Indole-3-Aldehyde In Bacteria Metabolic Network Of Tryptophan Around The Indole-3-Aldehyde Formation
Organism: Limosilactobacillus fermentum
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2023-07-05 Classification: OXIDOREDUCTASE Ligands: HEM |

