Search Count: 12
 |
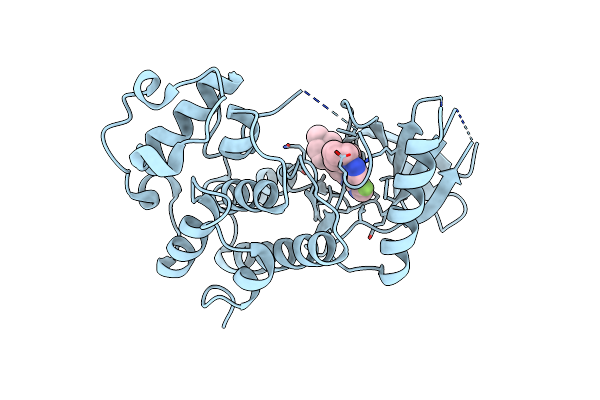
Cdk2 In Complex With Cpd14 (5-Fluoro-4-(4-Methyl-5,6,7,8-Tetrahydro-4H-Pyrazolo[1,5-A]Azepin-3-Yl)-N-(5-(4-Methylpiperazin-1-Yl)Pyridin-2-Yl)Pyrimidin-2-Amine)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-07-29 Classification: TRANSFERASE/TRANSFERASE inhibitor Ligands: N14 |
 |
Cdk6 In Complex With Cpd13 (R)-5-Fluoro-4-(4-Methyl-5,6,7,8-Tetrahydro-4H-Pyrazolo[1,5-A]Azepin-3-Yl)-N-(5-(4-Methylpiperazin-1-Yl)Pyridin-2-Yl)Pyrimidin-2-Amine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2020-07-29 Classification: TRANSFERASE/TRANSFERASE inhibitor Ligands: N1A |
 |
Cdk6 In Complex With Cpd24 N-(5-(6-Ethyl-2,6-Diazaspiro[3.3]Heptan-2-Yl)Pyridin-2-Yl)-5-Fluoro-4-(4-Methyl-5,6,7,8-Tetrahydro-4H-Pyrazolo[1,5-A]Azepin-3-Yl)Pyrimidin-2-Amine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2020-07-29 Classification: TRANSFERASE/TRANSFERASE inhibitor Ligands: N1J |
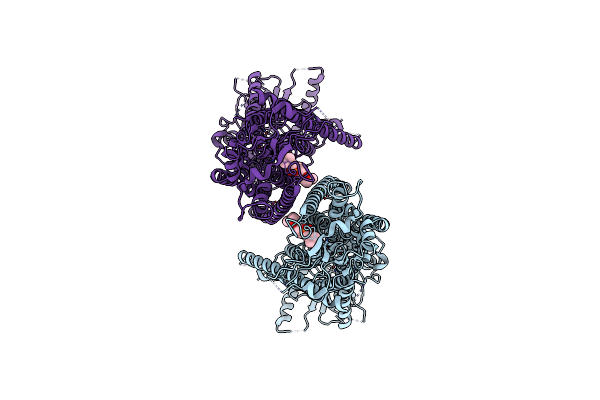 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2019-03-06 Classification: MEMBRANE PROTEIN Ligands: CA, P1O |
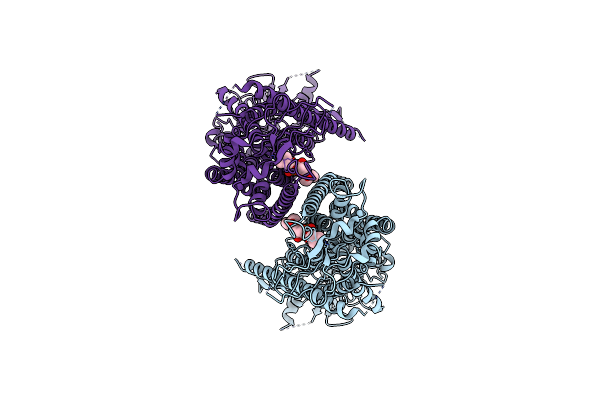 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2019-03-06 Classification: MEMBRANE PROTEIN Ligands: P1O |
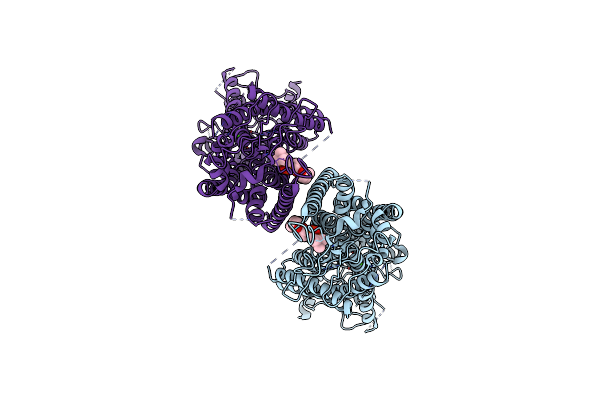 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2019-03-06 Classification: MEMBRANE PROTEIN Ligands: CA, P1O |
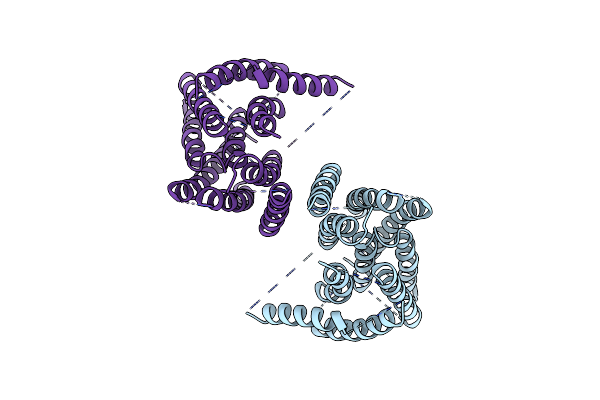 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2019-03-06 Classification: MEMBRANE PROTEIN |
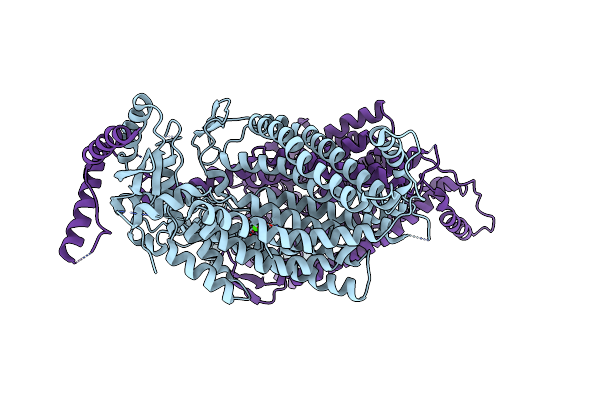 |
Organism: Nectria haematococca
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2014-11-12 Classification: LIPID TRANSPORT Ligands: CA |
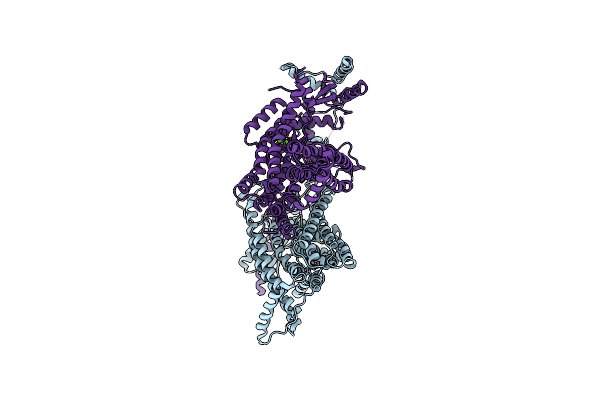 |
Organism: Nectria haematococca
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2014-11-12 Classification: LIPID TRANSPORT Ligands: CA |
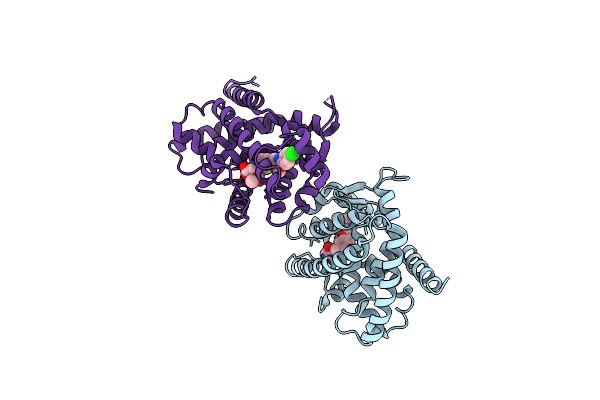 |
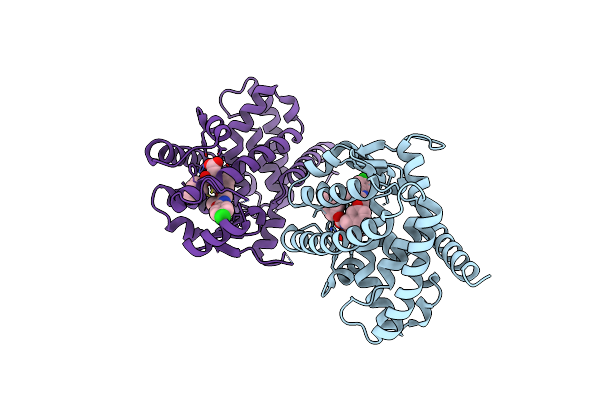
Crystal Structure Of Peroxisome Proliferator-Activatedeceptor Alpha (Pparalpha) Complex With N-3-((2-(4-Chlorophenyl)-5-Methyl-1,3-Oxazol-4-Yl)Methoxy)Benzyl)-N-(Methoxycarbonyl)Glycine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2010-04-28 Classification: HORMONE RECEPTOR Ligands: 7HA |
 |
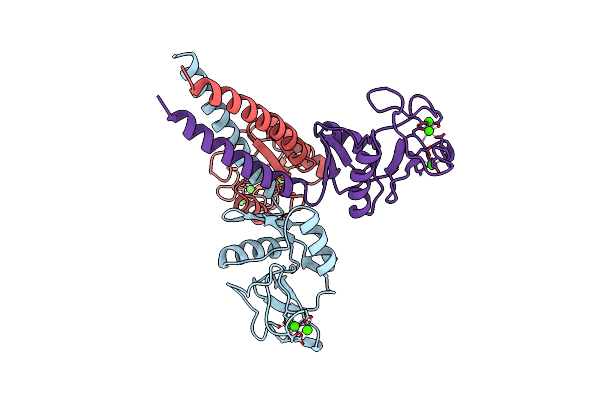
Crystal Structure Of Peroxisome Proliferator-Activatedeceptor Alpha (Pparalpha) Complex With N-3-((2-(4-Chlorophenyl)-5-Methyl-1,3-Oxazol-4-Yl)Methoxy)Benzyl)-N-((4-Methylphenoxy)Carbonyl)Glycine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2010-04-28 Classification: HORMONE RECEPTOR Ligands: NKS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 1999-12-01 Classification: SUGAR BINDING PROTEIN Ligands: CA |

