Search Count: 7
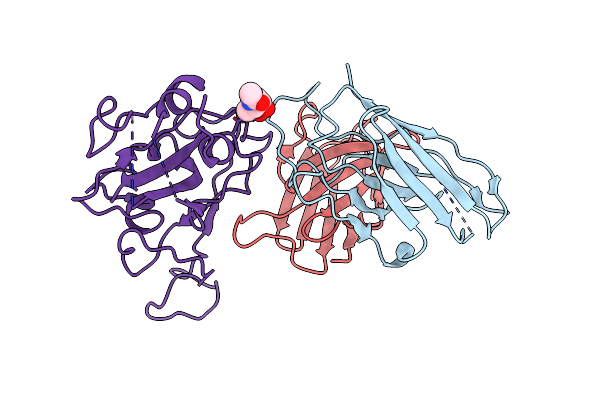 |
Cryo-Em Map Of Sars-Cov-2 Omicron Ba.2 Spike In Complex With 2130-1-0114-112
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus
Method: ELECTRON MICROSCOPY Release Date: 2024-02-21 Classification: IMMUNE SYSTEM Ligands: NAG |
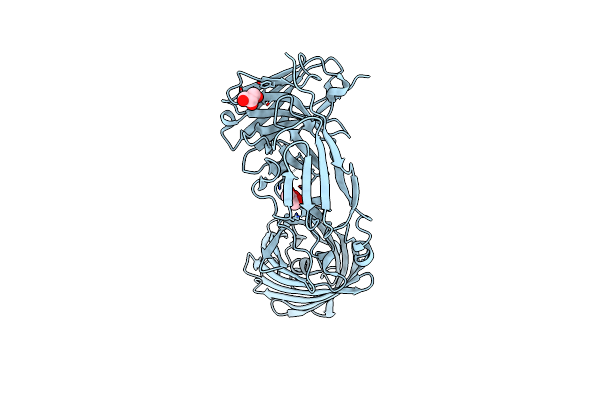 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-01-27 Classification: FLUORESCENT PROTEIN Ligands: GOL |
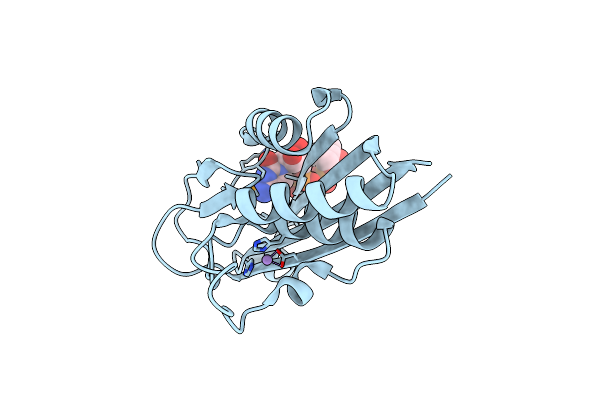 |
Co-Crystal Structure Of 2-C-Methyl-D-Erythritol 2,4-Cyclodiphosphate Synthase (Ispf) From E. Coli Involved In Mevalonate-Independent Isoprenoid Biosynthesis, Complexed With Cmp/Mecdp/Mn2+
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2002-06-18 Classification: METAL BINDING PROTEIN Ligands: MN, C5P, CDI |
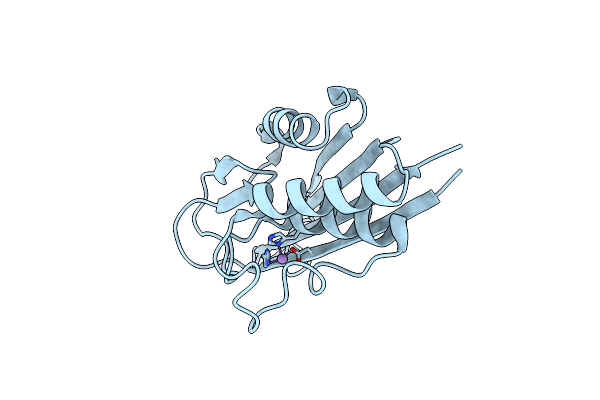 |
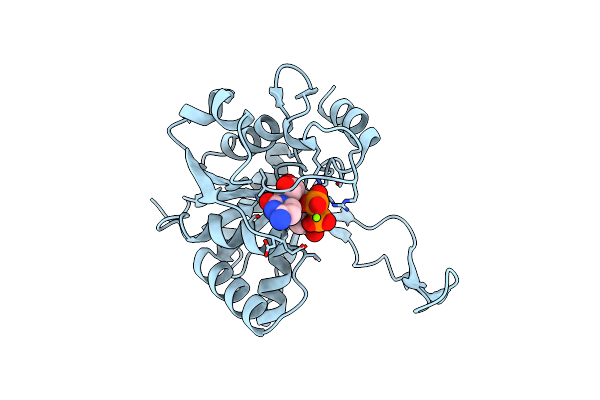
Crystal Structure Of 2-C-Methyl-D-Erythritol 2,4-Cyclodiphosphate Synthase (Ispf) From E. Coli Involved In Mevalonate-Independent Isoprenoid Biosynthesis
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2002-06-18 Classification: METAL BINDING PROTEIN Ligands: MN |
 |
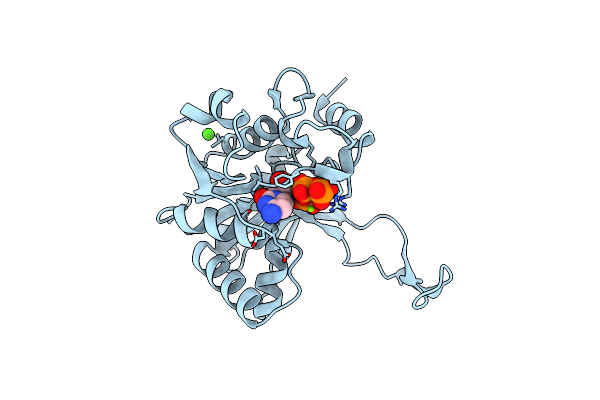
Crystal Structure Of 4-Diphosphocytidyl-2-C-Methylerythritol (Cdp-Me) Synthetase (Ygbp) Involved In Mevalonate Independent Isoprenoid Biosynthesis, Complexed With Cdp-Me And Mg2+
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2001-11-14 Classification: TRANSFERASE Ligands: MG, CDM |
 |
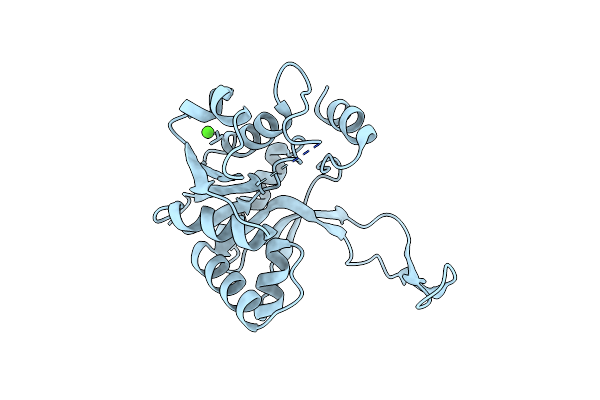
Crystal Structure Of 4-Diphosphocytidyl-2-C-Methylerythritol (Cdp-Me) Synthase (Ygbp) Involved In Mevalonate Independent Isoprenoid Biosynthesis
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2001-07-11 Classification: TRANSFERASE Ligands: CA, MG, CTP |
 |
Crystal Structure Of The Apo Form Of 4-Diphosphocytidyl-2-C-Methylerythritol (Cdp-Me) Synthetase (Ygbp) Involved In Mevalonate Independent Isoprenoid Biosynthesis
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2001-07-11 Classification: TRANSFERASE Ligands: CA |

