Search Count: 25
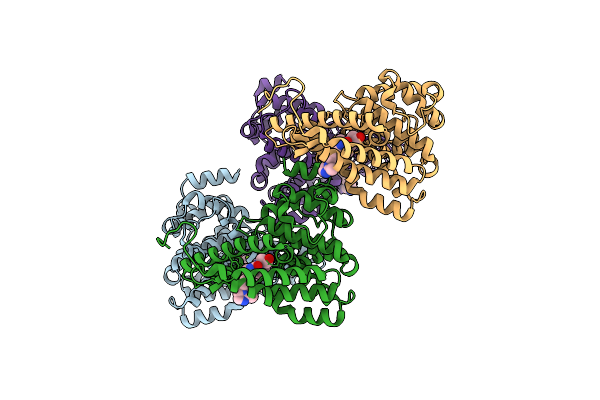 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2013-05-01 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: ZN, MG, 1L6 |
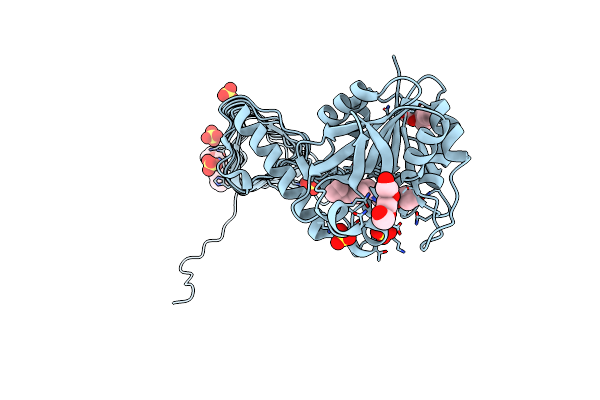 |
Crystal Structure Of Glmu From Haemophilus Influenzae In Complex With Quinazoline Inhibitor 1
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2009-11-17 Classification: TRANSFERASE Ligands: LZR, PG4, PGE, SO4 |
 |
Crystal Structure Of Glmu From Haemophilus Influenzae In Complex With Quinazoline Inhibitor 2
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2009-11-17 Classification: TRANSFERASE Ligands: LZS, PG4 |
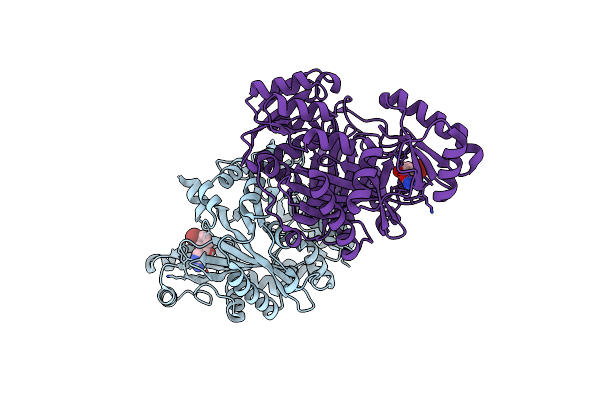 |
Crystal Structure Of Biotin Carboxylase From E. Coli In Complex With Amino-Oxazole Fragment Series
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-05-19 Classification: LIGASE Ligands: OA1, CL |
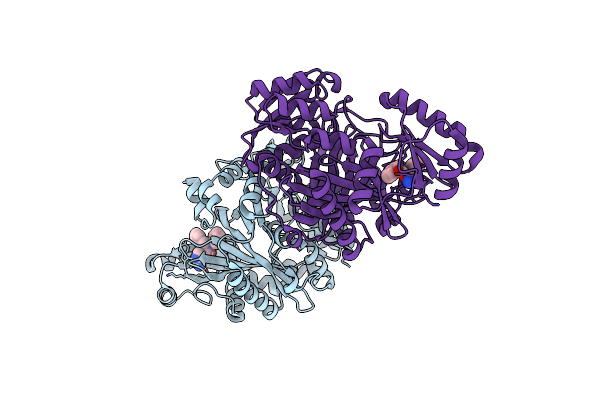 |
Crystal Structure Of Biotin Carboxylase From E. Coli In Complex With Amino-Oxazole Fragment Series
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2009-05-19 Classification: LIGASE Ligands: OA2, CL |
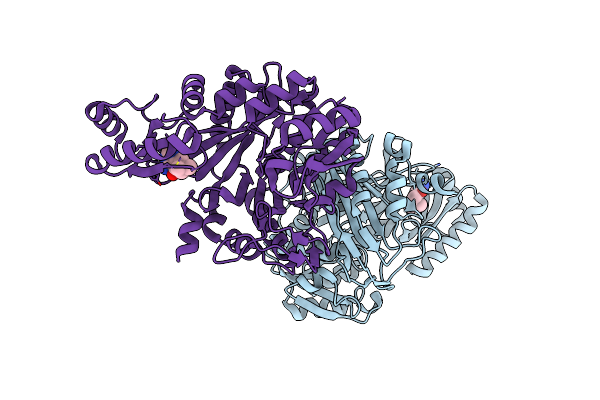 |
Crystal Structure Of Biotin Carboxylase From E. Coli In Complex With 4-Amino-7,7-Dimethyl-7,8-Dihydro-Quinazolinone Fragment
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2009-05-19 Classification: LIGASE Ligands: OA3, CL |
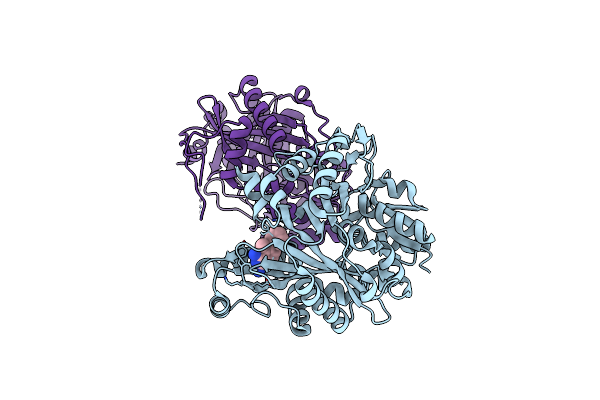 |
Crystal Structure Of Biotin Carboxylase From E. Coli In Complex With 5-Methyl-6-Phenyl-Quinazoline-2,4-Diamine
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2009-05-19 Classification: LIGASE Ligands: OA4 |
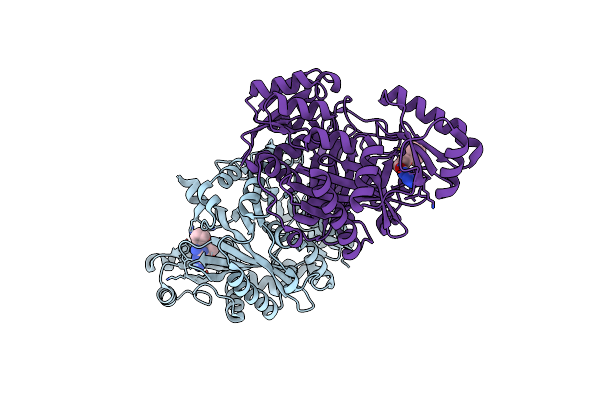 |
Crystal Structure Of Biotin Carboxylase From E. Coli In Complex With The Triazine-2,4-Diamine Fragment
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2009-05-19 Classification: LIGASE Ligands: OA5, CL |
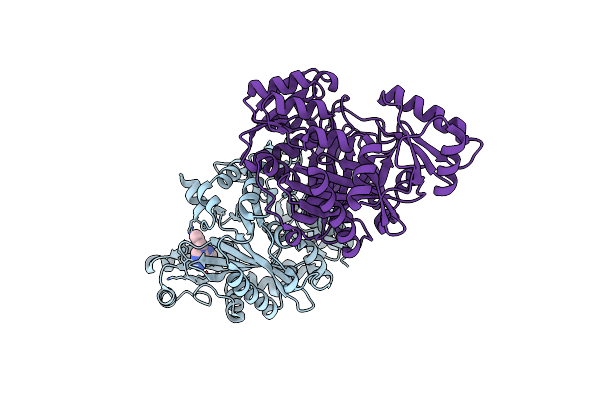 |
Crystal Structure Of Biotin Carboxylase From E. Coli In Complex With The 3-(3-Methyl-But-2-Enyl)-3H-Purin-6-Ylamine Fragment
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2009-05-19 Classification: LIGASE Ligands: L21, CL |
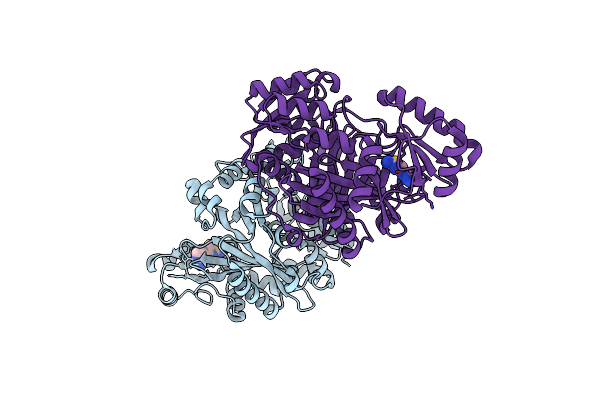 |
Crystal Structure Of Biotin Carboxylase From E. Coli In Complex With The Amino-Thiazole-Pyrimidine Fragment
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2009-05-19 Classification: LIGASE Ligands: L22, CL |
 |
Crystal Structure Of Biotin Carboxylase From E. Coli In Complex With The Imidazole-Pyrimidine Inhibitor
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2009-05-19 Classification: LIGASE Ligands: CL, L23 |
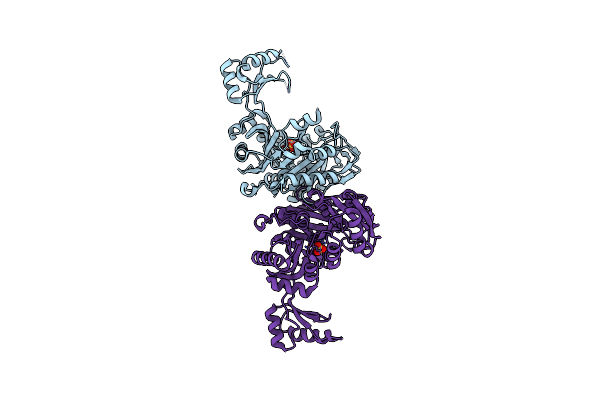 |
Crystal Structure Of Biotin Carboxylase From Pseudomonas Aeruginosa In Apo Form
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2008-09-09 Classification: LIGASE Ligands: SO4 |
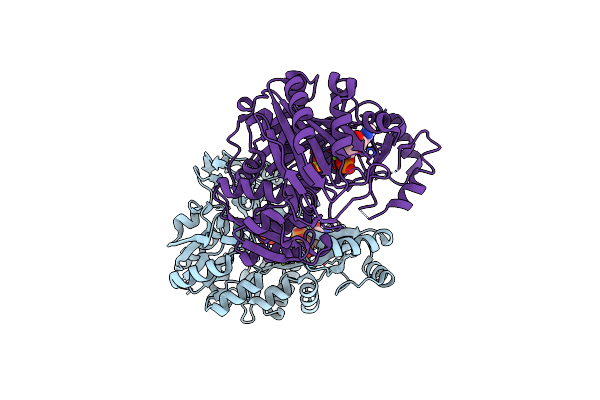 |
Crystal Structure Of Biotin Carboxylase From E. Coli In Complex With Amppnp And Adp
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2008-09-09 Classification: LIGASE Ligands: MG, SO4, ANP, ADP |
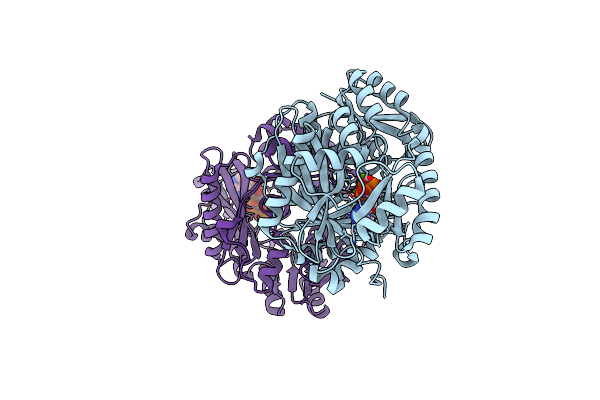 |
Crystal Structure Of Biotin Carboxylase From S. Aureus Complexed With Amppnp
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2008-09-09 Classification: LIGASE Ligands: ANP, MG, CL |
 |
Crystal Structure Of Biotin Carboxylase From Pseudomonas Aeruginosa Complexed With Ampcp
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2008-09-09 Classification: LIGASE Ligands: AP2, MG, SO4 |
 |
Crystal Structure Of Biotin Carboxylase From E. Coli In Complex With Atp Analog, Adpcf2P.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2008-09-09 Classification: LIGASE Ligands: CL, ATF |
 |
Characterization Of Substrate Binding And Catalysis Of The Potential Antibacterial Target N-Acetylglucosamine-1-Phosphate Uridyltransferase (Glmu)
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2008-01-15 Classification: TRANSFERASE Ligands: PEG, SO4, CO |
 |
Characterization Of Substrate Binding And Catalysis Of The Potential Antibacterial Target N-Acetylglucosamine-1-Phosphate Uridyltransferase (Glmu)
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2008-01-15 Classification: TRANSFERASE Ligands: UD1, PG4, PEG, SO4 |
 |
Characterization Of Substrate Binding And Catalysis Of The Potential Antibacterial Target N-Acetylglucosamine-1-Phosphate Uridyltransferase (Glmu)
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2008-01-15 Classification: TRANSFERASE Ligands: H2U, MG, PG4, PGE, SO4 |
 |
Characterization Of Substrate Binding And Catalysis Of The Potential Antibacterial Target N-Acetylglucosamine-1-Phosphate Uridyltransferase (Glmu)
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2008-01-15 Classification: TRANSFERASE Ligands: UDP, PG4, PGE, SO4 |

