Search Count: 30,480
 |
Cryo-Em Structure Of Rnf20/Rnf40-Rad6A-Ub In Complex With H2Bs112Glcnac Nucleosome
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: NUCLEAR PROTEIN Ligands: ZN, NAG |
 |
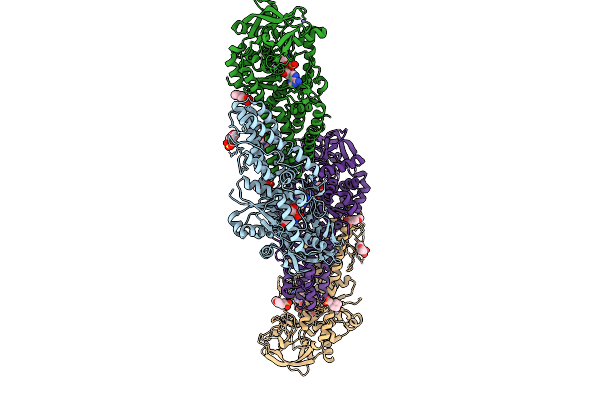
Fadd-Ded Filaments Coordinate Complex Iia Assembly During Tnf-Induced Apoptosis
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: IMMUNE SYSTEM |
 |
Organism: Streptomyces candidus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: LIGASE Ligands: MES, ZN, GSU |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: LIGASE Ligands: DMS, AMP |
 |
Dna Ligase 1 E346A/E592A In Complex With Nick Containing 3'-8Oxorg:A Captured At Post-Catalytic Stage
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: LIGASE/DNA Ligands: AMP |
 |
Dna Ligase 1 E346A/E592A In Complex With Nick Containing 3'-8Oxorg:C Captured At Pre-Catalytic Stage
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: LIGASE/DNA |
 |
Dna Ligase 1 E346A/E592A In Complex With Nick Containing 3'-8Oxodg:A Captured At Pre-Catalytic Stage
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: LIGASE/DNA Ligands: DMS |
 |
Dna Ligase 1 E346A/E592A In Complex With Nick Containing 3'-8Oxodg:C Captured At Pre-Catalytic Stage
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: LIGASE/DNA |
 |
Dna Ligase 1 Wild-Type In Complex With Nick Containing 3'-8Oxodg:C Captured At Pre-Catalytic Stage
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: LIGASE/DNA |
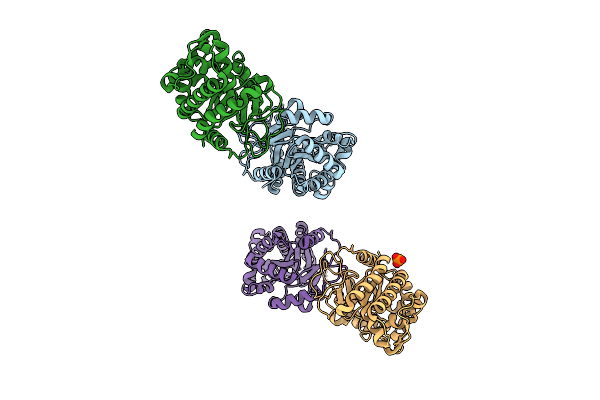 |
Organism: Thauera aromatica
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: TRANSFERASE Ligands: PO4 |
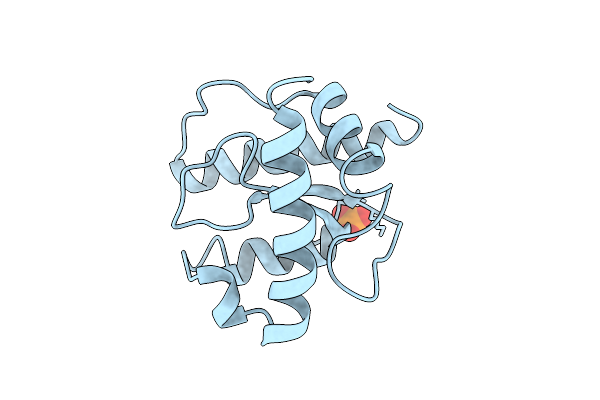 |
X-Ray Structure Of The C-Terminal Domain (Residues 366-485) Of S. Pombe Threonylcarbamoyladenosine Dehydratase
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: RNA BINDING PROTEIN Ligands: PO4 |
 |
X-Ray Structure Of S. Cerevisiae Threonylcarbamoyladenosine Dehydratase 1 (Residues 50-429) In Complex With Amp
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: RNA BINDING PROTEIN Ligands: AMP, CL, NA, GOL |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: LIGASE/ANTIBIOTIC Ligands: XMP, A1L67 |
 |
 |
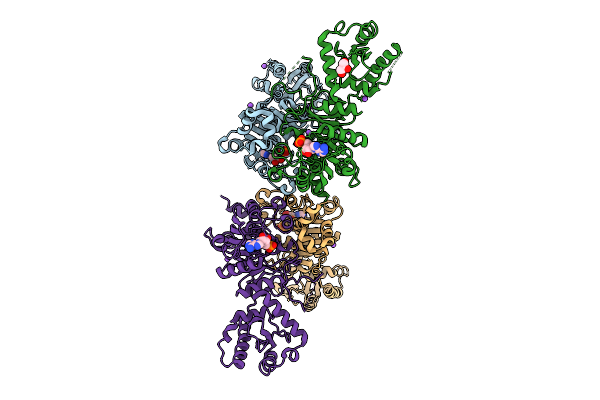
Structure Of Rbr Binding E2 Variant Crosslinked With Nedd8-Cul5-Rbx2 Bound Arih2 And Ub
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: LIGASE Ligands: ZN, SY8 |
 |
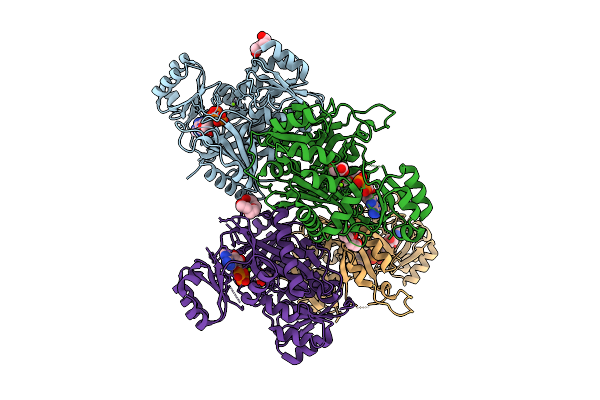
Organism: Synthetic construct, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: LIGASE Ligands: ZN |
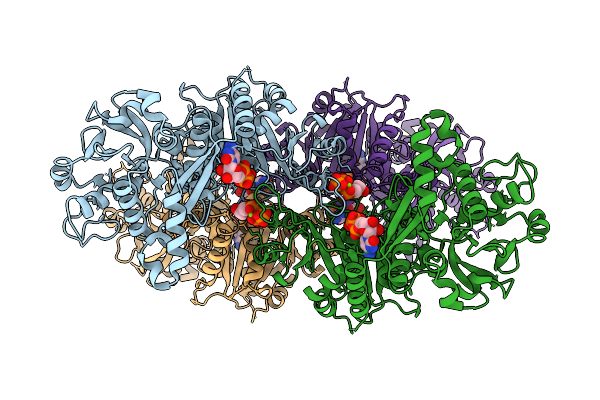 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: STRUCTURAL PROTEIN Ligands: CTP, MG |
 |
Crystal Structure Of Phosphoribosylaminoimidazole Carboxylase From Burkholderia Xenovorans (Atp Complex)
Organism: Paraburkholderia xenovorans lb400
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: LYASE Ligands: ATP, PEG, MG, PGE, GOL, ADP, PG4 |
 |
Crystal Structure Of Phosphoribosylaminoimidazole Carboxylase From Burkholderia Xenovorans (Adp Complex)
Organism: Paraburkholderia xenovorans lb400
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: LYASE Ligands: ADP, PEG, MG, PGE, GOL, PG4 |
 |
Crystal Structure Of Phosphoribosylaminoimidazole Carboxylase From Burkholderia Xenovorans (Amp Complex)
Organism: Paraburkholderia xenovorans lb400
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: LYASE Ligands: AMP, PGE, MG, PEG, PG4, GOL |

