Search Count: 24
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In Its Pfr State (I0A).
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL, EDO |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In Its Pfr State (I0B).
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4 |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I1 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4 |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I2 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, PGE, PEG, CL |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I3 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, GOL, PEG |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I4 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL, PEG |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I5 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL |
 |
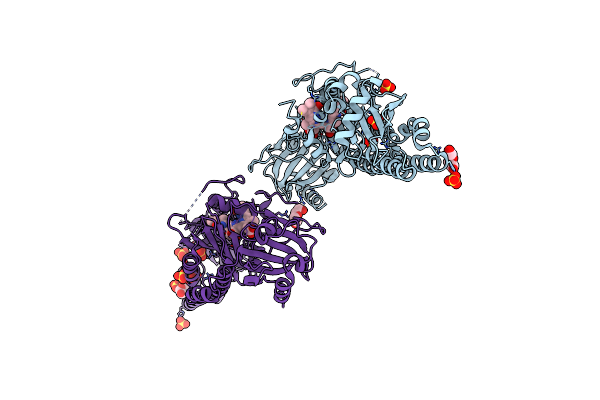
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I6 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I7 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, PEG |
 |
Alpha-N-Acetylgalactosaminidase Nagbb From Bifidobacterium Bifidum - Galnac Complex
Organism: Bifidobacterium bifidum
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-06-07 Classification: HYDROLASE Ligands: ZN, A2G, CA |
 |
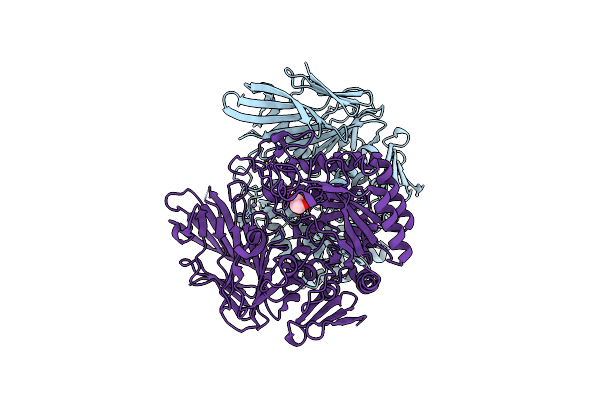
Alpha-N-Acetylgalactosaminidase Nagbb From Bifidobacterium Bifidum - Ligand Free
Organism: Bifidobacterium bifidum
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2017-06-07 Classification: HYDROLASE Ligands: ZN, CA |
 |
Alpha-N-Acetylgalactosaminidase Nagbb From Bifidobacterium Bifidum - Quadruple Mutant
Organism: Bifidobacterium bifidum
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-06-07 Classification: HYDROLASE Ligands: ZN, GOL |
 |
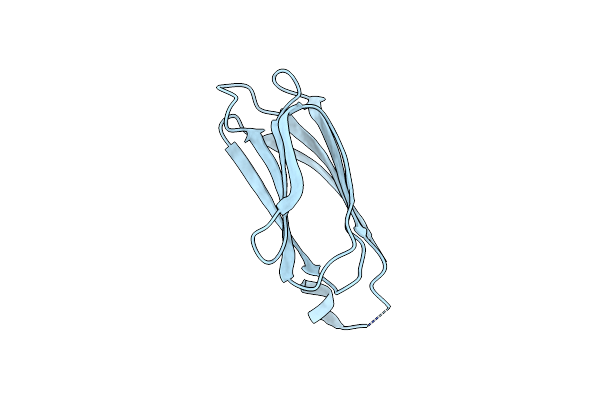
Alpha-N-Acetylgalactosaminidase Nagbb From Bifidobacterium Bifidum - Gal-Nhac-Dnj Complex
Organism: Bifidobacterium bifidum
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2017-06-07 Classification: HYDROLASE Ligands: ZN, DJN, CA |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2017-01-25 Classification: TRANSPORT PROTEIN Ligands: BR |
 |
Organism: Thermosynechococcus elongatus (strain bp-1)
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2016-11-23 Classification: ELECTRON TRANSPORT Ligands: OEX, FE2, SQD, CL, CLA, PHO, BCR, PL9, LMG, UNL, LHG, BCT, DGD, HEM, HEC |
 |
Nh3-Bound Rt Xfel Structure Of Photosystem Ii 500 Ms After The 2Nd Illumination (2F) At 2.8 A Resolution
Organism: Thermosynechococcus elongatus (strain bp-1)
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2016-11-23 Classification: ELECTRON TRANSPORT Ligands: OEX, FE2, LMG, CL, CLA, PHO, BCR, PL9, SQD, UNL, BCT, LHG, DGD, HEM, HEC |
 |
Room Temperature Xfel Structure Of The Native, Doubly-Illuminated Photosystem Ii Complex
Organism: Thermosynechococcus elongatus (strain bp-1), Thermosynechococcus elongatus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2016-11-23 Classification: PHOTOSYNTHESIS Ligands: OEX, FE2, CL, BCT, CLA, PHO, BCR, PL9, SQD, LHG, UNL, LMG, DGD, HEM, HEC |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:0.80 Å Release Date: 2012-12-19 Classification: HYDROLASE Ligands: CA, BEN, SO4, GOL |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:0.75 Å Release Date: 2012-12-19 Classification: HYDROLASE Ligands: CA, BEN, SO4, GOL |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:0.87 Å Release Date: 2012-12-19 Classification: HYDROLASE Ligands: CA, BEN, SO4, GOL |

