Search Count: 17
 |
Organism: Homo sapiens, Lama glama, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: DE NOVO PROTEIN/IMMUNE SYSTEM |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: IMMUNE SYSTEM |
 |
Crystal Structure Of Designed Allosteric Facilitated Dissociation Switch As1
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: DE NOVO PROTEIN Ligands: CL |
 |
Crystal Structure Of Designed Allosteric Facilitated Dissociation Switch As1 In Complex State He
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: DE NOVO PROTEIN Ligands: PGO |
 |
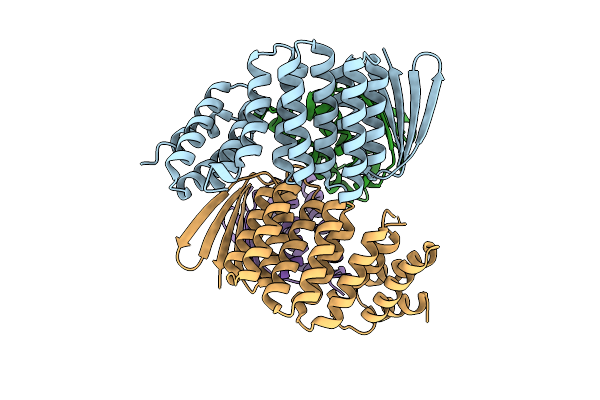
Designed Allosteric Facilitated Dissociation Switch As1 In Complex State Th With Methylated Lysines, Crystal #1
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: DE NOVO PROTEIN Ligands: MPD |
 |
Designed Allosteric Facilitated Dissociation Switch As1 In Complex State Th With Methylated Lysines, Crystal #2
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: DE NOVO PROTEIN |
 |
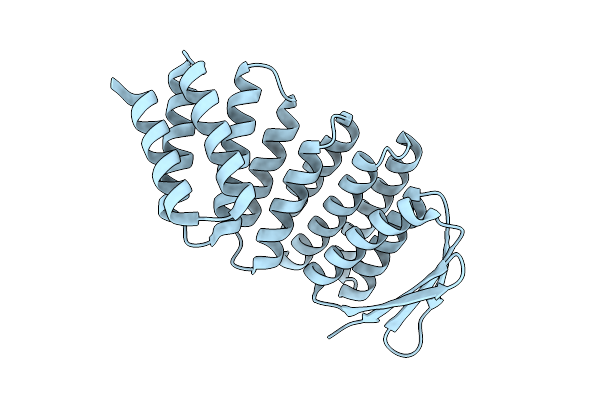
Designed Allosteric Facilitated Dissociation Switch As1 In Complex State The With Methylated Lysines
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: DE NOVO PROTEIN |
 |
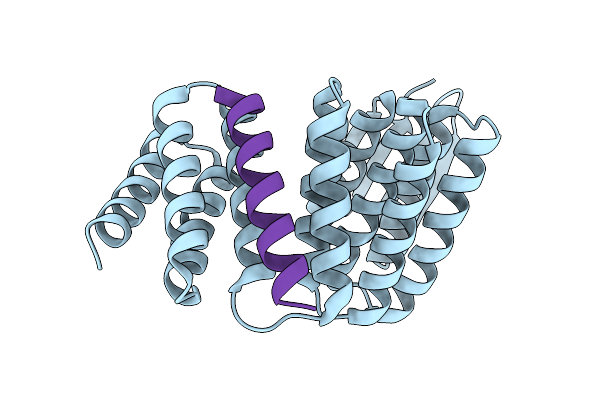
Crystal Structure Of Designed Allosteric Facilitated Dissociation Switch As5
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of Designed Allosteric Facilitated Dissociation Switch As5 In Complex State He
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: DE NOVO PROTEIN |
 |
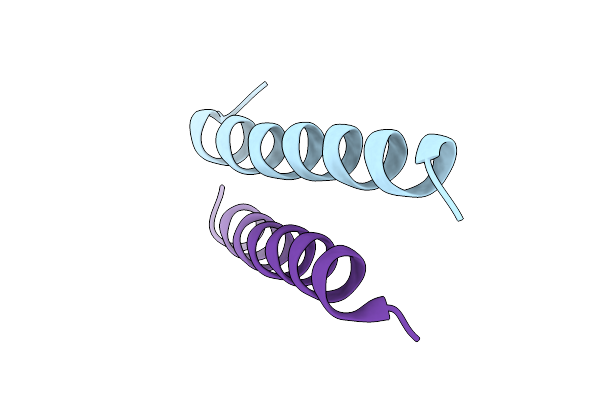
Crystal Structure Of Designed Facilitated Dissociation Target Lhd101An1: Lhd101A With An N-Terminal Extension
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of Designed Conformational Switch Effector Peptide Cs221B
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: DE NOVO PROTEIN |
 |
Designed Allosteric Facilitated Dissociation Switch As1_K46L_E50W_K172W_E173Y In Complex State The
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of Hiv-1 Reverse Transcriptase (Rt) In Complex With Janssen-R147681
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2004-05-11 Classification: TRANSFERASE Ligands: TPB |
 |
Crystal Structure Of Hiv-1 Reverse Transcriptase (Rt) In Complex With Janssen-R129385
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2004-05-11 Classification: TRANSFERASE Ligands: ADB |
 |
Crystal Structure Of Hiv-1 Reverse Transcriptase (Rt) In Complex With Janssen-R120394.
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2004-05-11 Classification: TRANSFERASE Ligands: ABZ |
 |
Crystal Structure Of Hiv-1 Reverse Transcriptase (Rt) In Complex With Janssen-R185545
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2004-05-11 Classification: TRANSFERASE Ligands: MG, 185 |
 |
Crystal Structure Of K103N Mutant Hiv-1 Reverse Transcriptase (Rt) In Complex With Janssen-R165335
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2004-05-11 Classification: TRANSFERASE Ligands: 65B |

