Search Count: 83
 |
Organism: Streptomyces graminofaciens, Synthetic construt
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: TRANSFERASE |
 |
Class 3 State Of The Gfsa Ksq-Ancestralat Chimeric Didomain In Complex With The Gfsa Acp Domain
Organism: Streptomyces graminofaciens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: LYASE Ligands: 9EF |
 |
Class 1 State Of The Gfsa Ksq-Ancestralat Chimeric Didomain In Complex With The Gfsa Acp Domain
Organism: Streptomyces graminofaciens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: LYASE Ligands: 9EF |
 |
Crystal Structure Of Flt3 In Complex With A Pyrazinamide Macrocycle Derivative
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2024-12-11 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: ZX3 |
 |
Crystal Structure Of The Acvr1 (Alk2) Kinase Domain In Complex With Inhibitor Cdd-2789
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2024-11-27 Classification: TRANSFERASE Ligands: GOL, A1A29, PO4 |
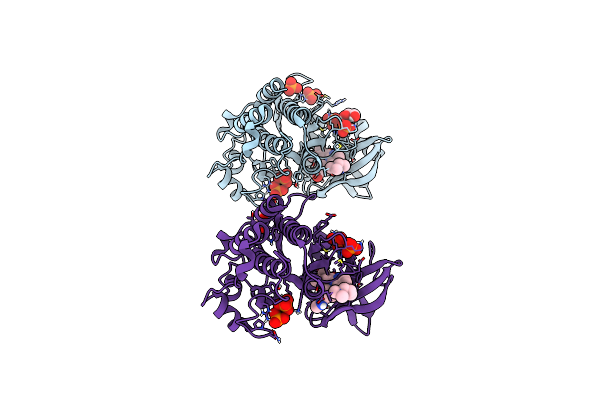 |
Crystal Structure Of The Acvr1 (Alk2) Kinase Domain In Complex With Inhibitor Cdd-2281
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2024-11-27 Classification: TRANSFERASE Ligands: PO4, GOL, A1A3C |
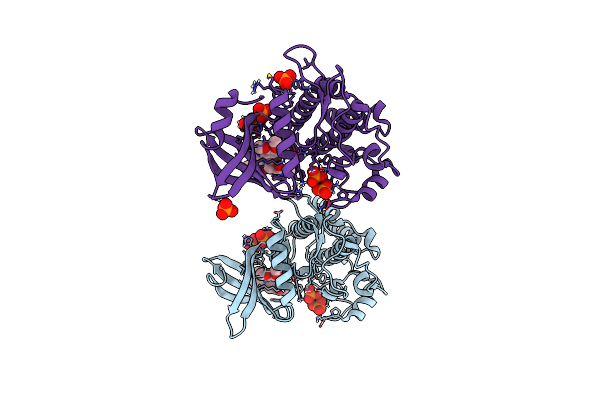 |
Crystal Structure Of The Acvr1 (Alk2) Kinase Domain In Complex With Inhibitor Cdd-2282
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-11-27 Classification: TRANSFERASE Ligands: A1A3D, PO4 |
 |
Organism: Saccharomonospora viridis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-11-06 Classification: HYDROLASE Ligands: NH4, BTB, C9C, GOL |
 |
Substrate Analog A010 Bound Form Of Pet-Degrading Cutinase Mutant Cut190**Ss_S176A
Organism: Saccharomonospora viridis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-11-06 Classification: HYDROLASE Ligands: A1L0N, CA |
 |
Substrate Analog A011 Bound Form Of Pet-Degrading Cutinase Mutant Cut190**Ss_S176A
Organism: Saccharomonospora viridis
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2024-11-06 Classification: HYDROLASE Ligands: A1L0L, CA |
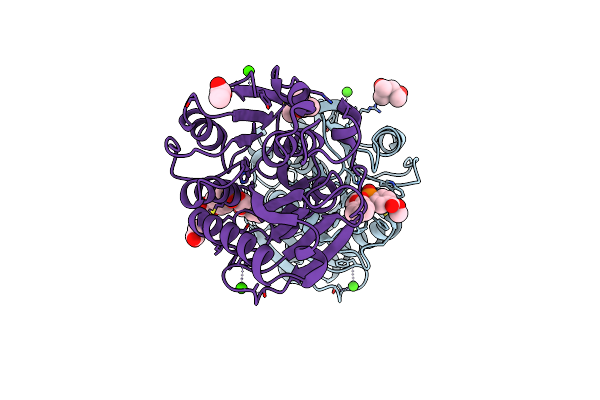 |
Substrate Analog A012 Bound Form Of Pet-Degrading Cutinase Mutant Cut190**Ss_S176A
Organism: Saccharomonospora viridis
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2024-11-06 Classification: HYDROLASE Ligands: MPD, A1L0M, CA, CL, ACT |
 |
Substrate Analog A013 Bound Form Of Pet-Degrading Cutinase Mutant Cut190**Ss_S176A
Organism: Saccharomonospora viridis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-11-06 Classification: HYDROLASE Ligands: A1L0O, CA |
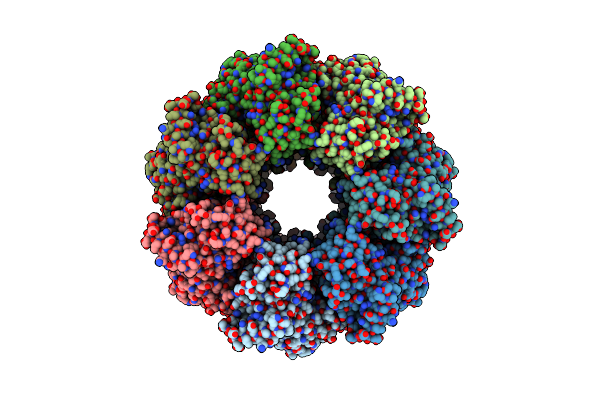 |
Organism: Hydrogenophilus thermoluteolus
Method: ELECTRON MICROSCOPY Release Date: 2024-03-27 Classification: CHAPERONE |
 |
Organism: Hydrogenophilus thermoluteolus
Method: ELECTRON MICROSCOPY Release Date: 2024-03-27 Classification: CHAPERONE Ligands: ADP, MG |
 |
Cryo-Em Structure Of H. Thermophilus Groel-Groes2 Asymmetric Football Complex
Organism: Hydrogenobacter thermophilus tk-6
Method: ELECTRON MICROSCOPY Release Date: 2024-03-27 Classification: CHAPERONE Ligands: MG, ANP, K |
 |
Organism: Hydrogenobacter thermophilus tk-6
Method: ELECTRON MICROSCOPY Release Date: 2024-03-27 Classification: CHAPERONE Ligands: MG, ANP, K |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-11-15 Classification: LIPID BINDING PROTEIN Ligands: MG, GTP |
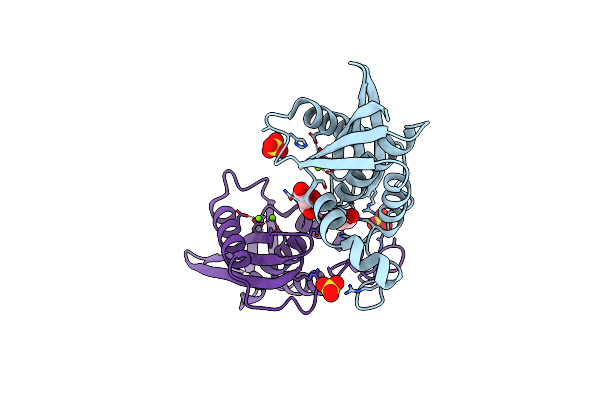 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2022-03-02 Classification: HYDROLASE Ligands: GOL, MG, SO4 |
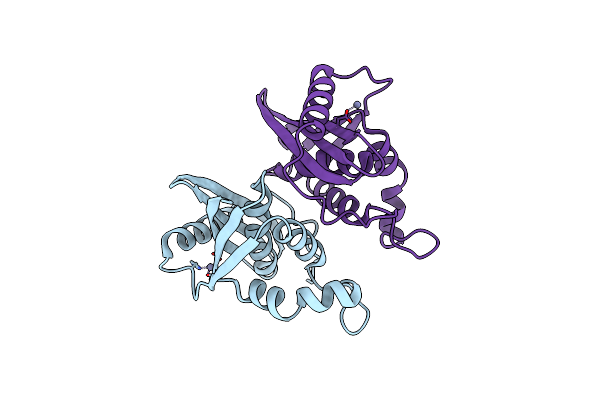 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2022-03-02 Classification: HYDROLASE Ligands: ZN |
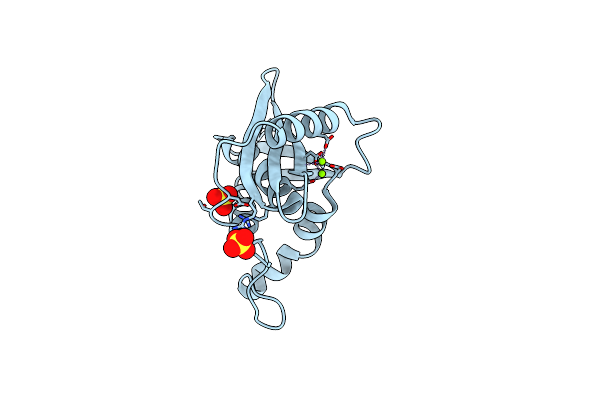 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2022-03-02 Classification: HYDROLASE Ligands: MG, SO4 |

