Search Count: 31
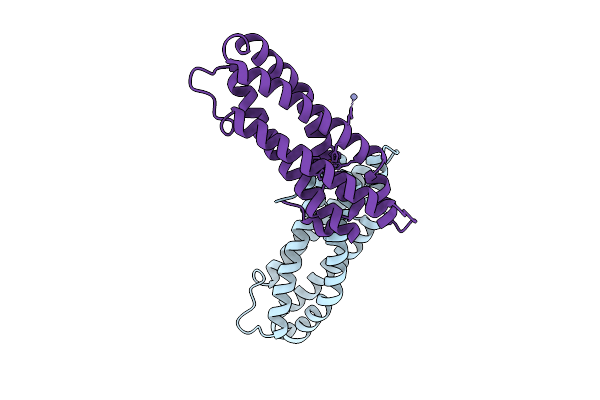 |
Crystal Structure Of Staphylococcus Aureus Cysteine-Free Scda With Bound Iron, Determined By Molecular Replacement And Fe Anomalous Signal
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: METAL BINDING PROTEIN Ligands: FE, O, ZN |
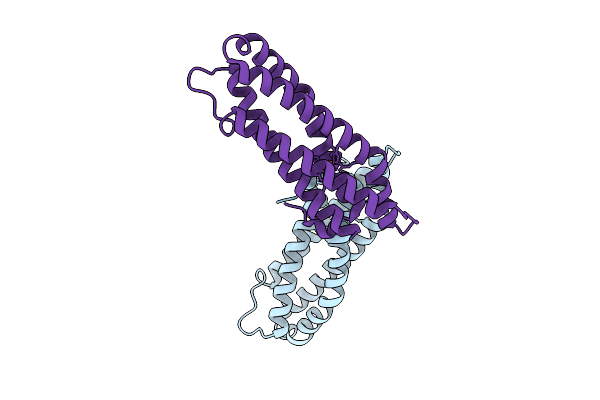 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: OXYGEN BINDING Ligands: FE, O |
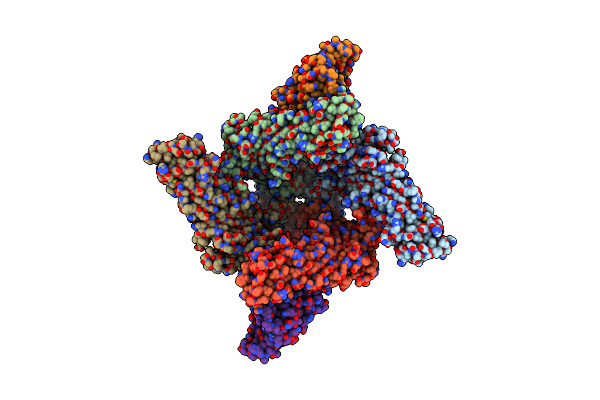 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: MEMBRANE PROTEIN Ligands: P5S, U6L, GDP, MG |
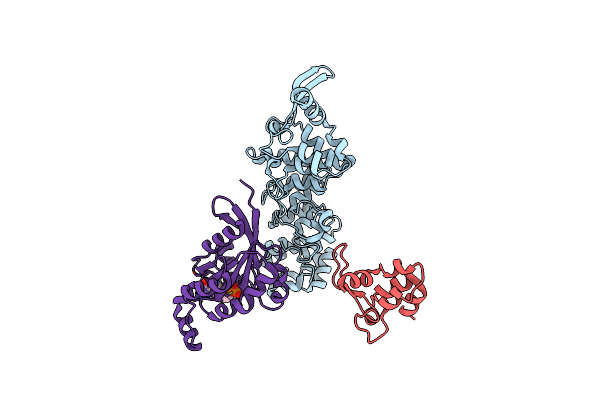 |
Cryo-Em Structure Of Human Trpv4 Intracellular Domain In Complex With Gtpase Rhoa
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: MEMBRANE PROTEIN Ligands: GDP, MG |
 |
Organism: Actinomadura sp.
Method: X-RAY DIFFRACTION Release Date: 2025-05-21 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Organism: Actinomadura sp.
Method: X-RAY DIFFRACTION Release Date: 2025-05-21 Classification: OXIDOREDUCTASE Ligands: HEM, A1EGF |
 |
Organism: Paenibacillus
Method: X-RAY DIFFRACTION Resolution:3.01 Å Release Date: 2025-03-19 Classification: RNA Ligands: GNG |
 |
Organism: Homo
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2025-03-19 Classification: RNA Ligands: SO4, NA |
 |
Organism: Homo
Method: X-RAY DIFFRACTION Resolution:3.86 Å Release Date: 2025-03-19 Classification: RNA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2025-03-19 Classification: RNA Ligands: MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2025-03-19 Classification: RNA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2025-03-19 Classification: RNA Ligands: GUN, BA, PO4, NA |
 |
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Release Date: 2025-02-26 Classification: RNA Ligands: GMP, NA |
 |
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Release Date: 2025-02-19 Classification: RNA Ligands: GTP, GNG, MG, NA |
 |
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Release Date: 2025-02-19 Classification: RNA Ligands: GTP, GUN, MG, NA |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2025-01-29 Classification: RNA Ligands: 2ZY, K, NA |
 |
Organism: Streptosporangium roseum
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-01-10 Classification: TRANSFERASE Ligands: PEG, ACT |
 |
Organism: Streptosporangium roseum, Saccharothrix mutabilis subsp. capreolus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-01-10 Classification: TRANSFERASE Ligands: ACT |
 |
Organism: Streptosporangium roseum, Saccharothrix mutabilis subsp. capreolus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-01-10 Classification: TRANSFERASE Ligands: ACT, PEG |
 |
Organism: Streptosporangium roseum
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2024-01-10 Classification: TRANSFERASE Ligands: PEG, ATP, ACT |

