Search Count: 220
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-12-17 Classification: DE NOVO PROTEIN Ligands: 308 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-12-17 Classification: DE NOVO PROTEIN Ligands: UNX, 308 |
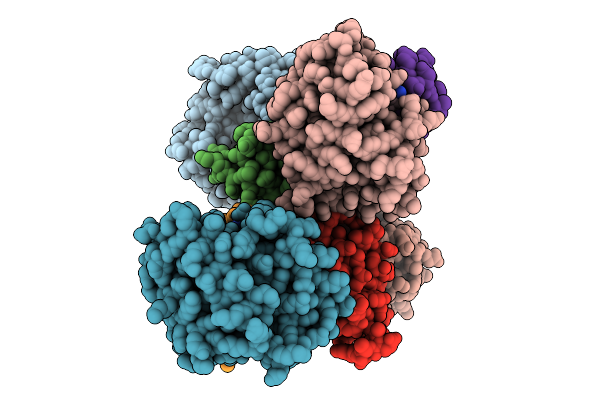 |
Crystal Structure Of De Novo Designed Amantadine Induced Heterodimer Daid23.4
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2025-12-17 Classification: DE NOVO PROTEIN Ligands: 308 |
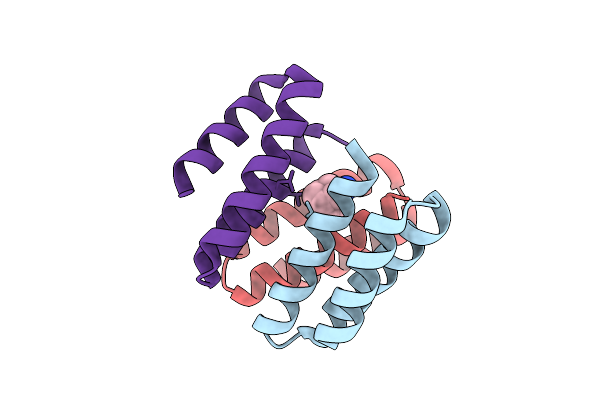 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-12-17 Classification: DE NOVO PROTEIN Ligands: 308 |
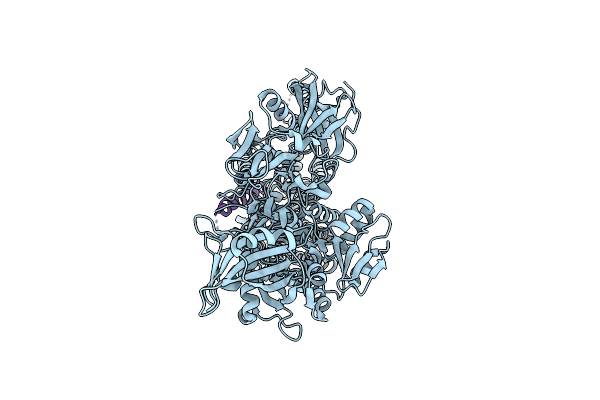 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.83 Å Release Date: 2025-06-11 Classification: MEMBRANE PROTEIN |
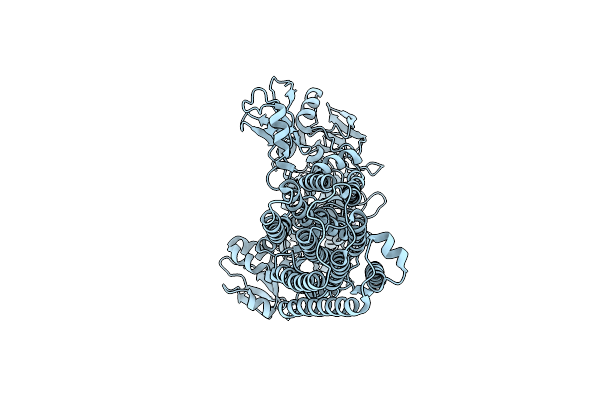 |
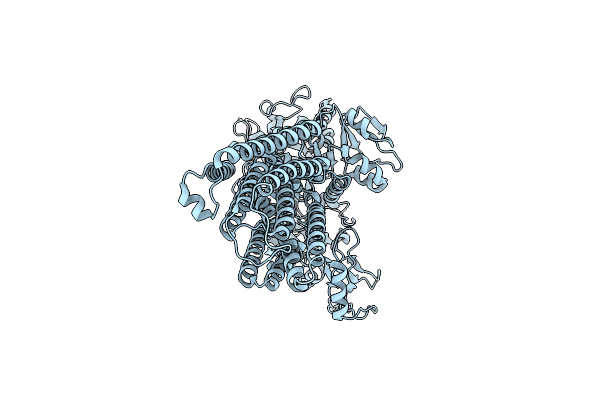
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.40 Å Release Date: 2025-06-11 Classification: MEMBRANE PROTEIN |
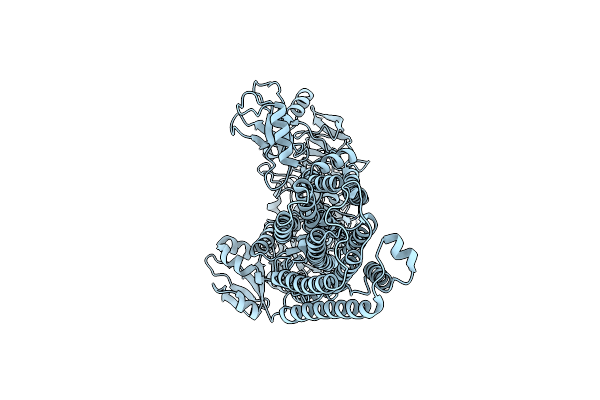 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.87 Å Release Date: 2025-06-11 Classification: MEMBRANE PROTEIN |
 |
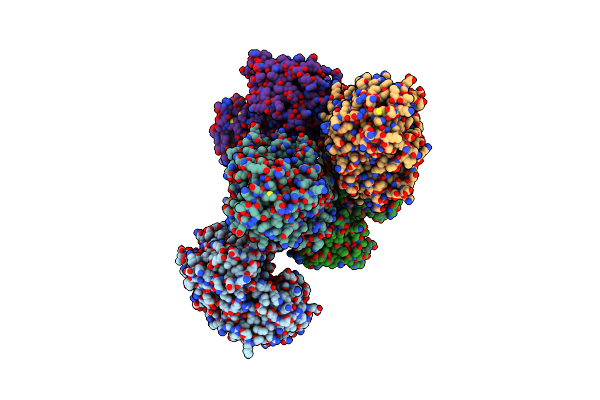
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: MEMBRANE PROTEIN |
 |
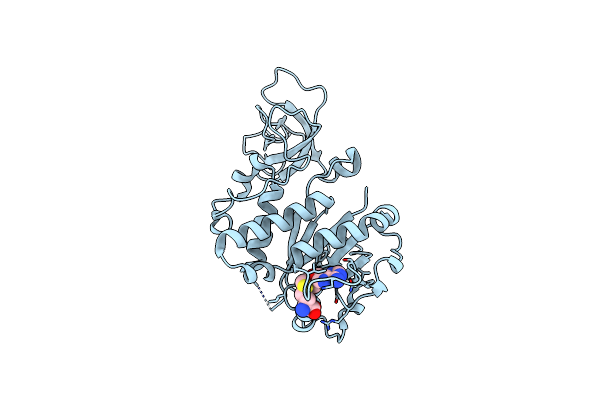
Glucosyl Transferase Nbugt72Ay1 Co-Crystallized With Scopoletin And Udp2Fglucose In The Presence Of Retinol
Organism: Nicotiana benthamiana
Method: X-RAY DIFFRACTION Resolution:3.11 Å Release Date: 2025-02-12 Classification: HYDROLASE Ligands: U2F, T83 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2024-10-23 Classification: CELL CYCLE Ligands: SAH |
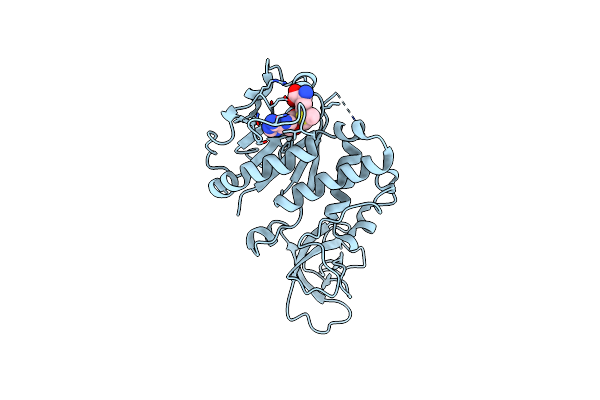 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2024-10-23 Classification: CELL CYCLE Ligands: SAM |
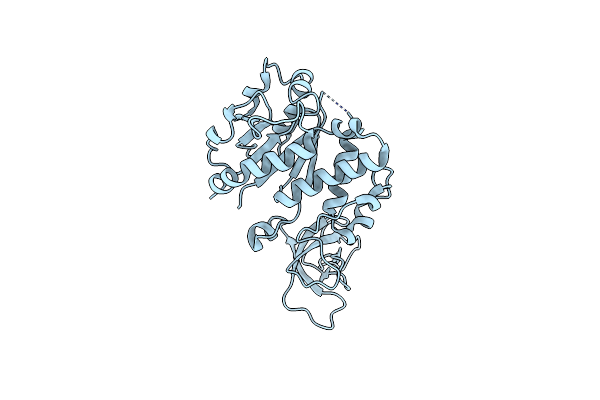 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-10-23 Classification: CELL CYCLE Ligands: HOH |
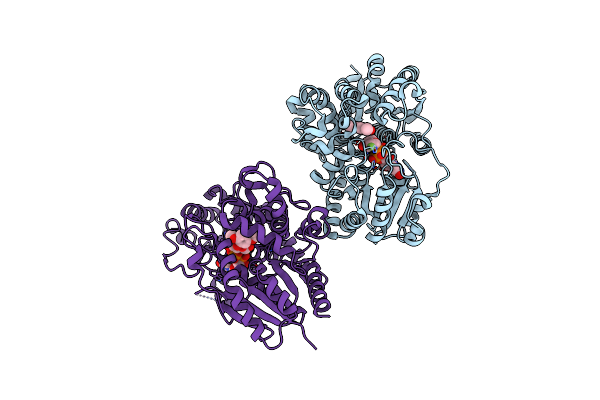 |
Glucosyl Transferase Nbugt72Ay1 Co-Crystallized With Scopoletin And Udp2F Glucose
Organism: Nicotiana benthamiana
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2024-09-04 Classification: TRANSFERASE Ligands: T83, U2F |
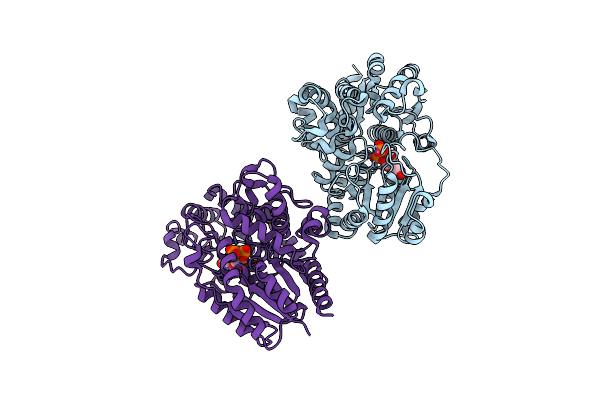 |
Organism: Nicotiana tabacum
Method: X-RAY DIFFRACTION Resolution:3.32 Å Release Date: 2024-04-17 Classification: TRANSFERASE Ligands: UDP |
 |
Organism: Nicotiana tabacum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-04-17 Classification: TRANSFERASE Ligands: T83, GOL |
 |
Organism: Nicotiana tabacum
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2024-04-17 Classification: TRANSFERASE |
 |
Organism: Nicotiana tabacum
Method: X-RAY DIFFRACTION Resolution:3.78 Å Release Date: 2024-04-17 Classification: TRANSFERASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2024-03-27 Classification: TRANSFERASE/INHIBITOR Ligands: A1LVQ, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-03-27 Classification: TRANSFERASE/INHIBITOR Ligands: A1LVQ, SO4 |
 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2024-02-28 Classification: TRANSFERASE Ligands: CLM, GOL, SO4 |

