Search Count: 19
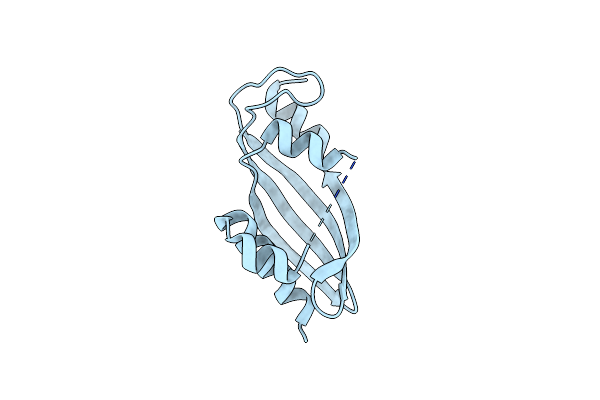 |
Crystal Structure Of Dyna1, A Putative Monoxygenase From Mivromonospora Chersina.
Organism: Micromonospora chersina
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-09-04 Classification: ISOMERASE |
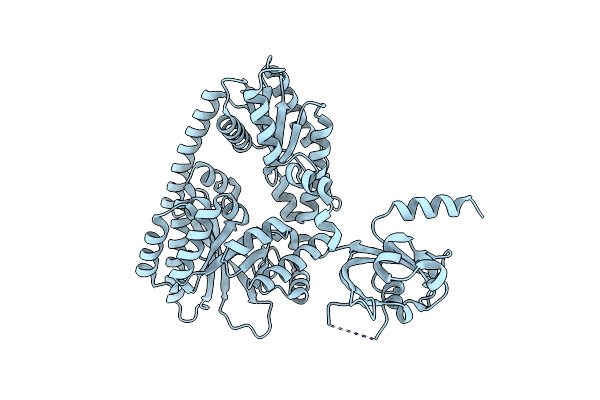 |
Crystal Structure Of The Pas-Ggdef-Eal Domain Of Pa0861 From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2017-12-20 Classification: TRANSCRIPTION |
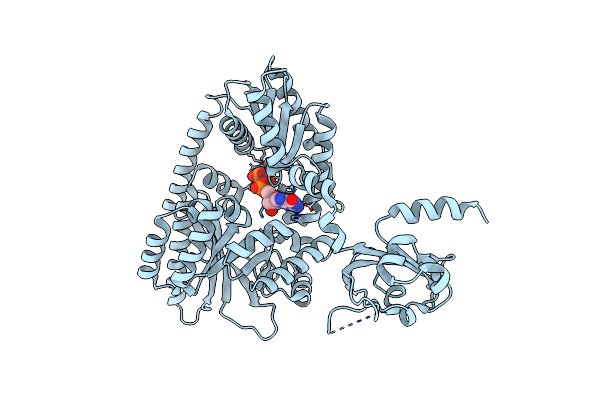 |
Crystal Structure Of The Pas-Ggdef-Eal Domain Of Pa0861 From Pseudomonas Aeruginosa In Complex With Gtp
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2017-12-20 Classification: TRANSCRIPTION Ligands: GTP, MG |
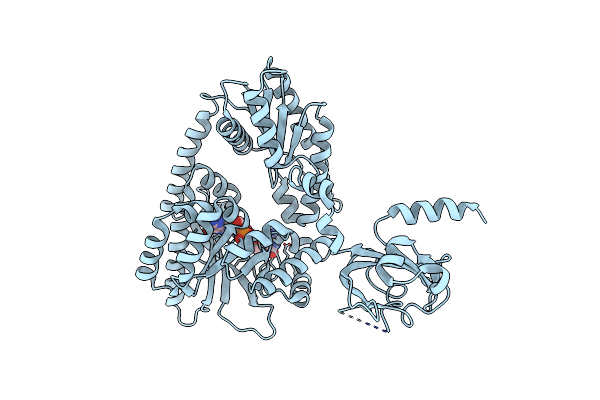 |
Crystal Structure Of The Pas-Ggdef-Eal Domain Of Pa0861 From Pseudomonas Aeruginosa In Complex With Cyclic Di-Gmp
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:3.31 Å Release Date: 2017-12-20 Classification: TRANSCRIPTION Ligands: C2E |
 |
Organism: Pseudomonas aeruginosa str. pao1
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-11-29 Classification: TRANSFERASE/PROTEIN BINDING Ligands: SO4, C2E |
 |
Organism: Pseudomonas aeruginosa str. pao1
Method: X-RAY DIFFRACTION Resolution:3.41 Å Release Date: 2017-11-29 Classification: TRANSFERASE |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2017-02-01 Classification: TRANSPORT PROTEIN Ligands: 2BA |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2015-10-07 Classification: IMMUNE SYSTEM Ligands: 2BA |
 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-10-08 Classification: LYASE Ligands: C2E |
 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2014-10-08 Classification: TRANSFERASE |
 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2014-10-08 Classification: HYDROLASE Ligands: GOL, MES, C2E |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2013-05-29 Classification: IMMUNE SYSTEM Ligands: C2E |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2013-05-29 Classification: IMMUNE SYSTEM |
 |
Organism: Geobacillus thermodenitrificans
Method: SOLUTION NMR Release Date: 2013-03-27 Classification: HYDROLASE |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-07-18 Classification: TRANSCRIPTION REGULATOR Ligands: MG, EPE |
 |
Organism: Micromonospora echinospora
Method: SOLUTION NMR Release Date: 2011-06-15 Classification: TRANSFERASE |
 |
Induced-Fit And Allosteric Effects Upon Polyene Binding Revealed By Crystal Structures Of The Dynemicin Thioesterase
Organism: Micromonospora chersina
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2010-10-13 Classification: BIOSYNTHETIC PROTEIN Ligands: SSV |
 |
Crystal Structure Of A Bifunctional Hotdog Fold Thioesterase In Enediyne Biosynthesis, Cale7
Organism: Micromonospora echinospora
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2009-04-07 Classification: HYDROLASE Ligands: PO4, JEF, POP, GOL |
 |
Crystal Structure Of Nad(+)-Dependent Alcohol Dehydrogenase From Bacillus Stearothermophilus Strain Lld-R
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2004-05-11 Classification: OXIDOREDUCTASE Ligands: ZN, ETF |

