Search Count: 84
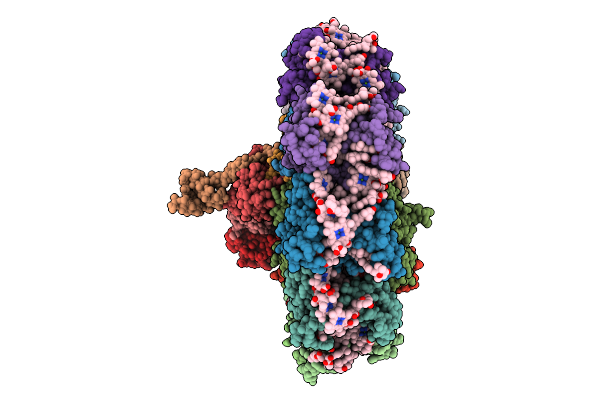 |
Organism: Tribonema minus
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: PHOTOSYNTHESIS Ligands: CLA, BCR, DD6, LMT, LHG, A1L1G, LMG, PQN, SF4 |
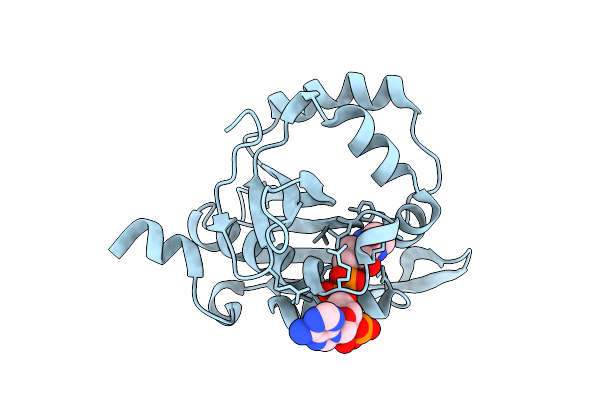 |
Iterative Acetyltransferase On Lasso Peptides From Actinomycetes In Complex With Coa
Organism: Actinosynnema mirum (strain atcc 29888 / dsm 43827 / jcm 3225 / nbrc 14064 / ncimb 13271 / nrrl b-12336 / imru 3971 / 101)
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: TRANSFERASE Ligands: COA |
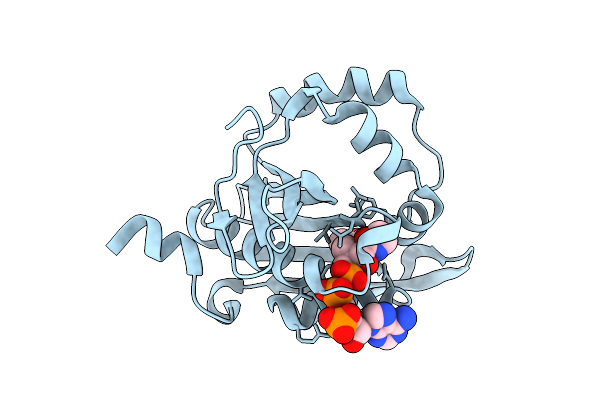 |
Iterative Acetyltransferase On Lasso Peptides From Actinomycetes In Complex With Accoa
Organism: Actinosynnema mirum (strain atcc 29888 / dsm 43827 / jcm 3225 / nbrc 14064 / ncimb 13271 / nrrl b-12336 / imru 3971 / 101)
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: TRANSFERASE Ligands: ACO |
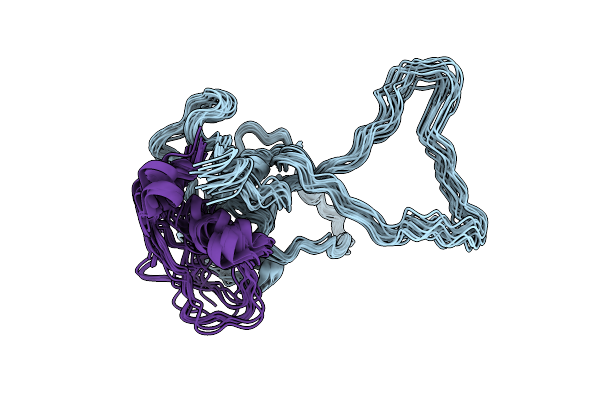 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2025-08-06 Classification: PROTEIN BINDING |
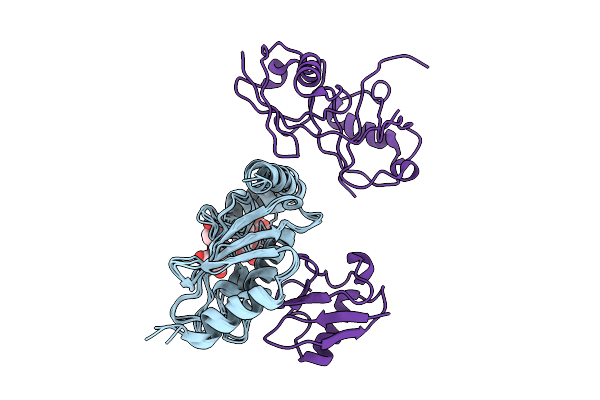 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2025-07-23 Classification: PROTEIN BINDING Ligands: A1EQF |
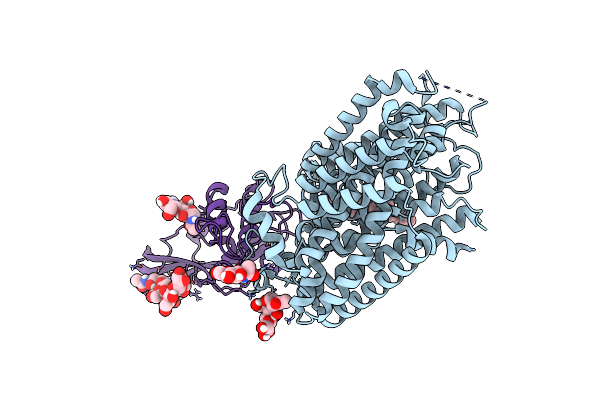 |
Cryo-Em Structure Of Mmcat1 Bound With Frmlv-Rbd In The Apo Inward-Open State
Organism: Mus musculus, Murine leukemia virus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: MEMBRANE PROTEIN Ligands: NAG, Y01 |
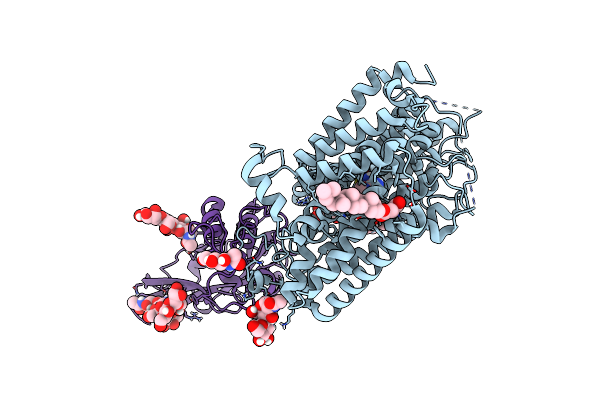 |
Cryo-Em Structure Of Mmcat1 Bound With Frmlv-Rbd In The Arginine-Bound Inward-Occluded State
Organism: Mus musculus, Murine leukemia virus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: MEMBRANE PROTEIN Ligands: NAG, Y01, ARG |
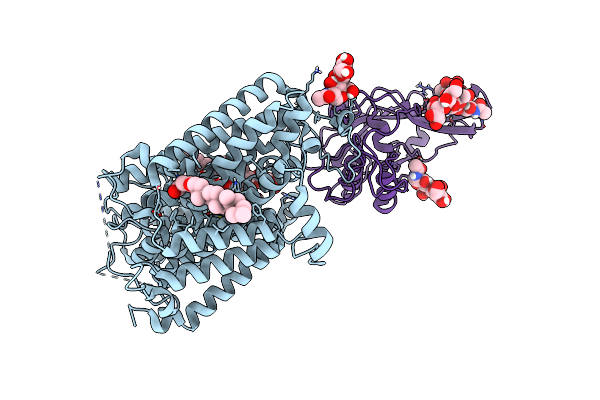 |
Cryo-Em Structure Of Mmcat1 Bound With Frmlv-Rbd In The Lysine-Bound Inward-Occluded State
Organism: Mus musculus, Murine leukemia virus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: MEMBRANE PROTEIN Ligands: LYS, Y01 |
 |
Cryo-Em Structure Of Mmcat1 Bound With Frmlv-Rbd In The Ornithine-Bound Inward-Occluded State
Organism: Mus musculus, Murine leukemia virus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: MEMBRANE PROTEIN Ligands: Y01, ORN |
 |
Cryo-Em Structure Of Enterovirus A71 Mature Virion In Complex With Fab Ct11F9
Organism: Mus musculus, Enterovirus a71
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: VIRUS Ligands: SPH |
 |
Organism: Bacteroides xylanisolvens xb1a
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2025-01-01 Classification: HYDROLASE Ligands: GOL |
 |
Organism: Bacteroides xylanisolvens xb1a
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2025-01-01 Classification: HYDROLASE Ligands: GLA |
 |
Beta-Galactosidase From Bacteroides Xylanisolvens (Complex With Methyl Beta-Galactopyranose)
Organism: Bacteroides xylanisolvens xb1a
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2025-01-01 Classification: HYDROLASE Ligands: MBG |
 |
Crystal Structure Of Dyna1, A Putative Monoxygenase From Mivromonospora Chersina.
Organism: Micromonospora chersina
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-09-04 Classification: ISOMERASE |
 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-05-08 Classification: HYDROLASE, IMMUNE SYSTEM |
 |
Crystal Structure Of Estzf172 As A Novel Biocatalyst For The Efficient Biosynthesis Of A Chiral Intermediate Of Pregabalin
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Release Date: 2024-05-01 Classification: LYASE |
 |
Crystal Structure Of Tr3 Lbd In Complex With Para-Positioned 3,4,5-Trisubstituted Benzene Derivatives
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-01-24 Classification: TRANSCRIPTION Ligands: XBF |
 |
Organism: Aquifex aeolicus vf5
Method: ELECTRON MICROSCOPY Release Date: 2023-09-06 Classification: MEMBRANE PROTEIN Ligands: HEM, POV |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: VIRAL PROTEIN/ANTIVIRAL PROTEIN Ligands: NAG |

