Search Count: 38
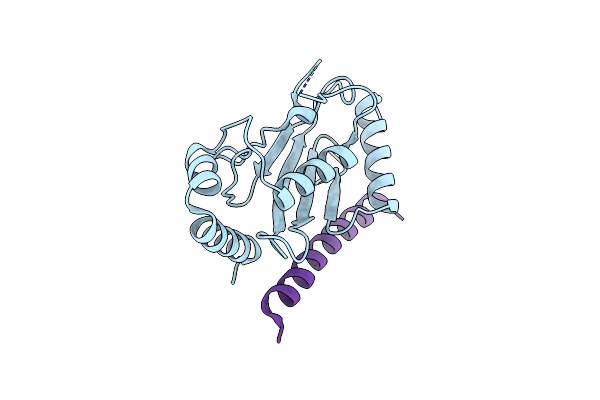 |
Crystal Structure Of Ube2G2 In Complex With The Ube2G2-Binding Region Of Aup1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2021-11-10 Classification: LIGASE/Transferase |
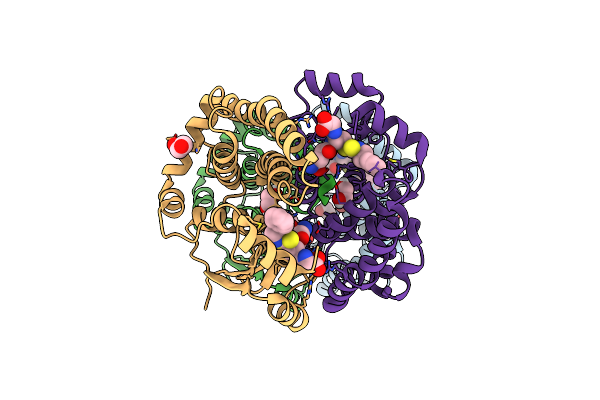 |
Crystal Structure Of Hgsta1-1 With Glutathione Adduct Of Phenethyl Isothiocyanate And Cystein Adduct Of Phenethyl Isothiocyanate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2016-10-12 Classification: TRANSFERASE Ligands: GVX, EDO |
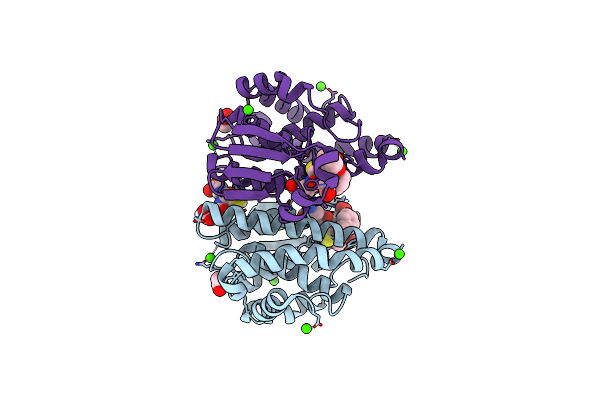 |
Crystal Structure Of Hgstp1-1 With Glutathione Adduct Of Phenethyl Isothiocyanate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2016-10-12 Classification: TRANSFERASE Ligands: GVX, MPD, ACT, CA, GSH |
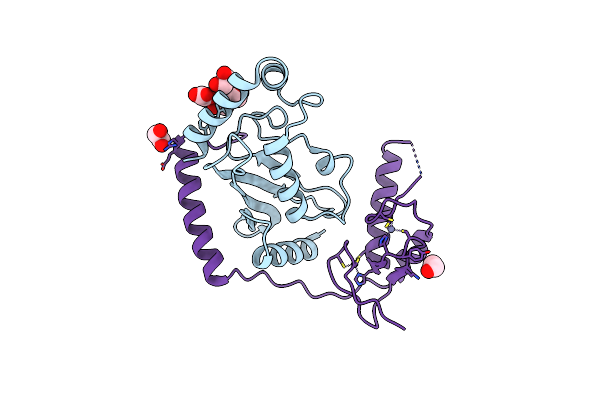 |
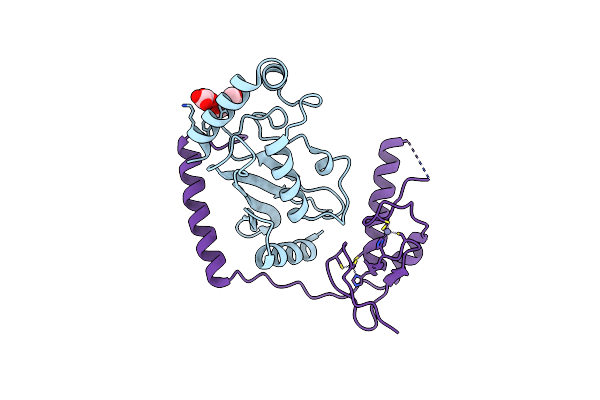
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2015-10-28 Classification: LIGASE Ligands: EDO, PEG, OXL, ZN |
 |
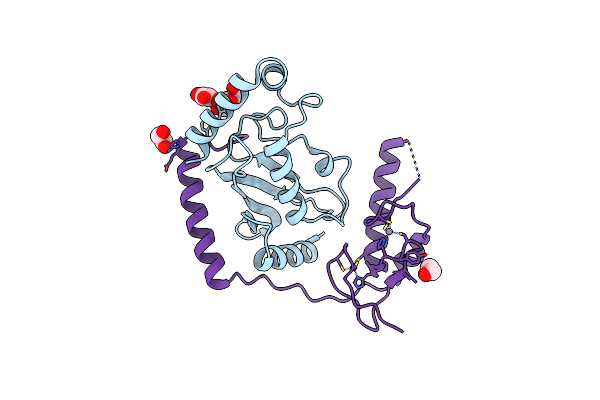
Crystal Structure Of Ubch5B In Complex With The Ring-U5Br Fragment Of Ao7 (Y165A)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2015-10-28 Classification: LIGASE Ligands: PEG, OXL, ZN |
 |
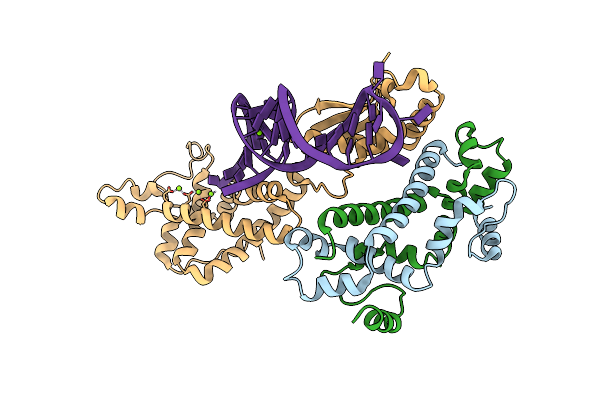
Crystal Structure Of Ubch5B In Complex With The Ring-U5Br Fragment Of Ao7 (P199A)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2015-10-28 Classification: LIGASE Ligands: OXL, PEG, ZN, EDO |
 |
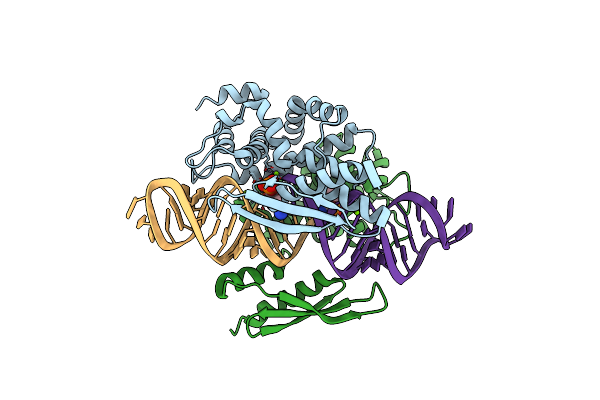
Crystal Structure Of Yeast Rnase Iii (Rnt1P) Complexed With The Product Of Dsrna Processing
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2014-04-16 Classification: HYDROLASE/RNA Ligands: MG |
 |
Crystal Structure Of Rnase Iii Complexed With Double-Stranded Rna And Cmp (Type Ii Cleavage)
Organism: Aquifex aeolicus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2013-12-11 Classification: HYDROLASE/RNA Ligands: MG, C5P |
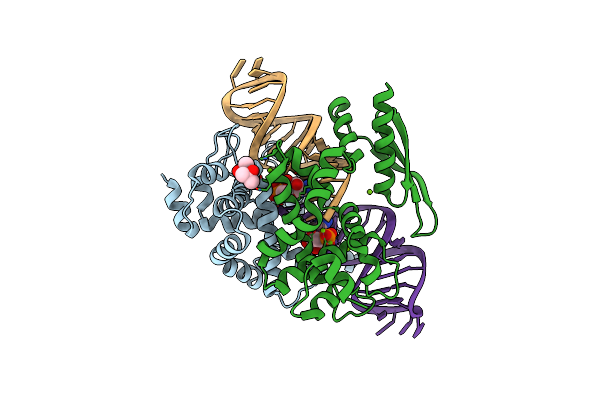 |
Crystal Structure Of Rnase Iii Complexed With Double-Stranded Rna And Amp (Type Ii Cleavage)
Organism: Aquifex aeolicus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2013-12-11 Classification: HYDROLASE/RNA Ligands: MG, MPD, AMP |
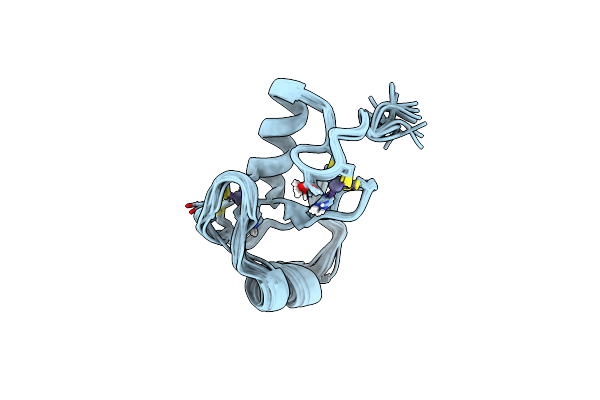 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2013-08-28 Classification: LIGASE Ligands: ZN |
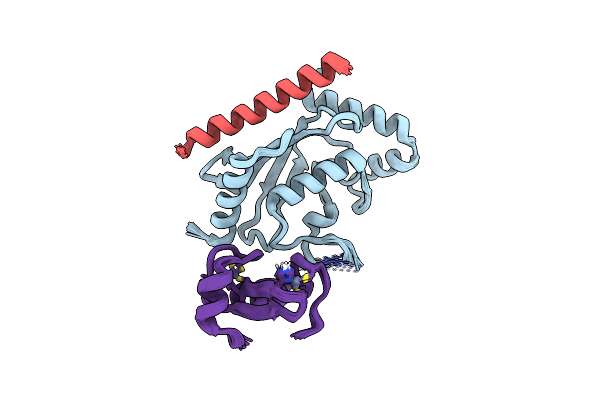 |
Nmr Structure Of Two Domains In Ubiquitin Ligase Gp78, Ring And G2Br, Bound To Its Conjugating Enzyme Ube2G
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2013-08-28 Classification: LIGASE Ligands: ZN |
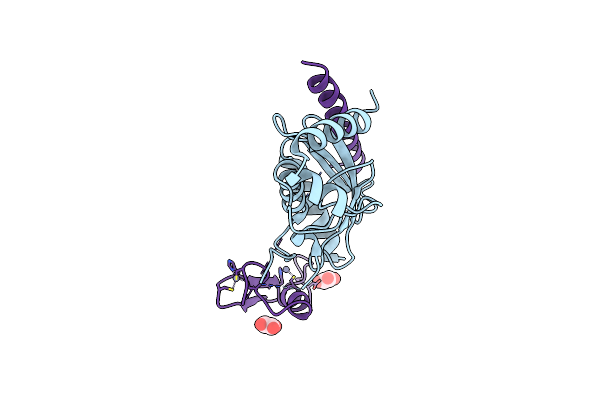 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-08-28 Classification: LIGASE/LIGASE Ligands: ZN, OXL |
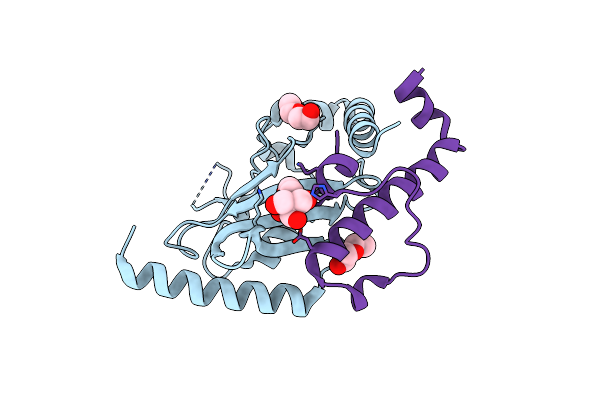 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2013-05-29 Classification: LIGASE/PROTEIN BINDING Ligands: BTB, PEG |
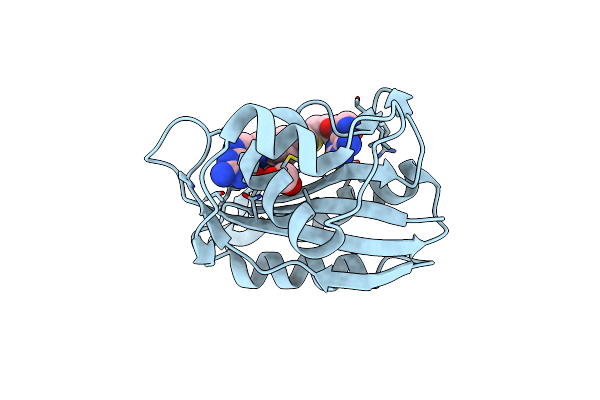 |
Crystal Structure Of E. Coli Hppk In Complex With Bisubstrate Analogue Inhibitor J1A
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-01-04 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: J1A, EDO |
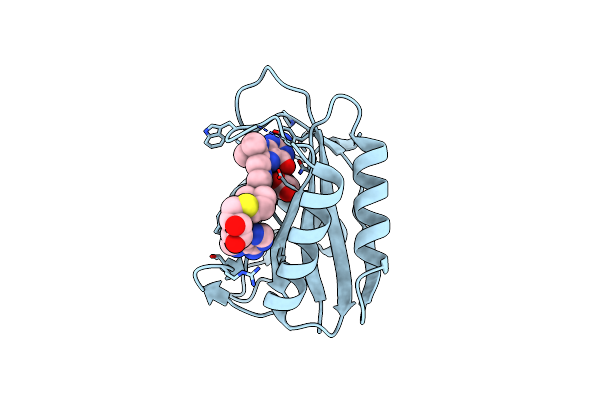 |
Crystal Structure Of E. Coli Hppk In Complex With Bisubstrate Analogue Inhibitor J1B
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2012-01-04 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: J1B, ACT |
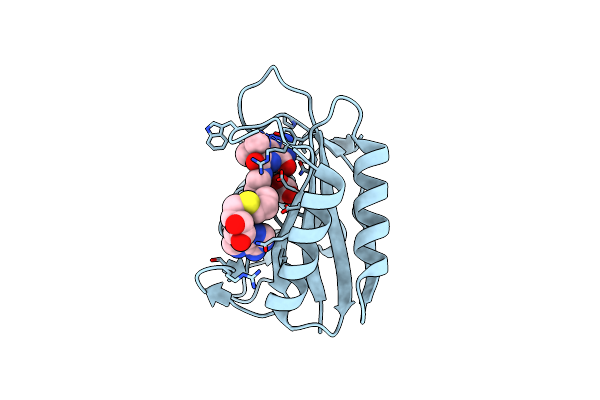 |
Crystal Structure Of E. Coli Hppk In Complex With Bisubstrate Analogue Inhibitor J1C
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2012-01-04 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: J1C, ACT |
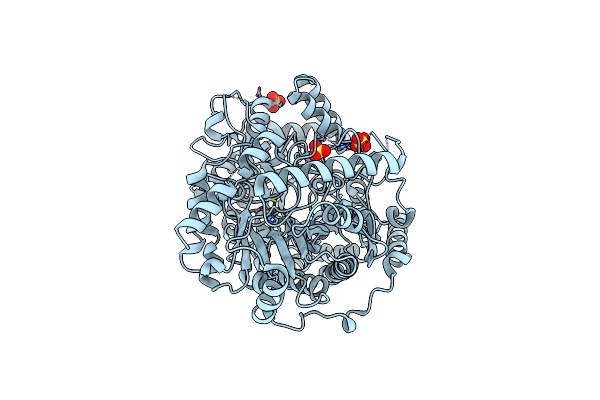 |
Organism: Streptococcus mutans
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2011-08-03 Classification: TRANSFERASE Ligands: SO4, ZN |
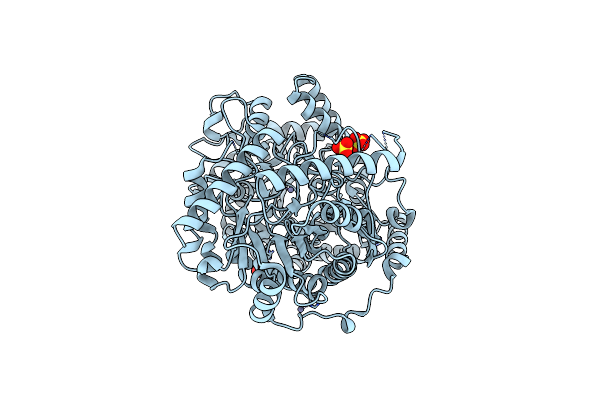 |
Organism: Streptococcus mutans
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2011-01-12 Classification: TRANSFERASE Ligands: SO4, ZN |
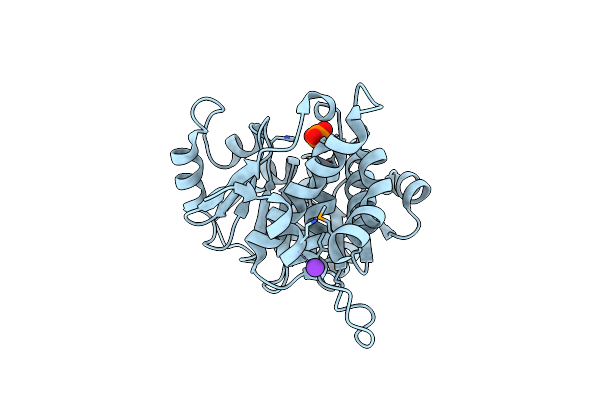 |
The Crystal Structure Of Hypothetic Protein Smu.573 From Streptococcus Mutans
Organism: Streptococcus mutans
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2008-12-16 Classification: UNKNOWN FUNCTION Ligands: PO4, K |
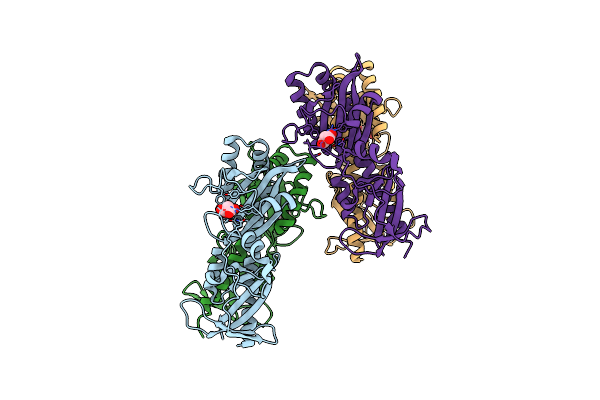 |
Organism: Rhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2008-11-18 Classification: TRANSFERASE Ligands: MLA |

