Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: ISOMERASE/INHIBITOR Ligands: CL |
 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2025-04-16 Classification: CELL CYCLE Ligands: A1A1H |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2025-04-16 Classification: CELL CYCLE Ligands: A1A1H, PEG, GOL, EDO |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: CELL CYCLE Ligands: ZN, A1A1I |
 |
Cryo-Em Structure Of Cdk2/Cycline1 In Complex With Crbn/Ddb1 And Cpd 4 (Local Mask)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: CELL CYCLE Ligands: ZN, A1A1I |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.27 Å Release Date: 2025-03-19 Classification: TRANSCRIPTION Ligands: A1AAL, EDO |
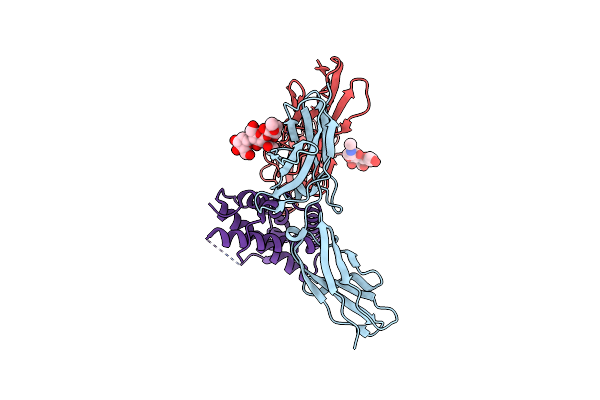 |
Structure Of The Ifn-Lambda4/Ifn-Lambdar1/Il-10Rbeta Receptor Complex With An Engineered Il-10Rbeta
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: CYTOKINE |
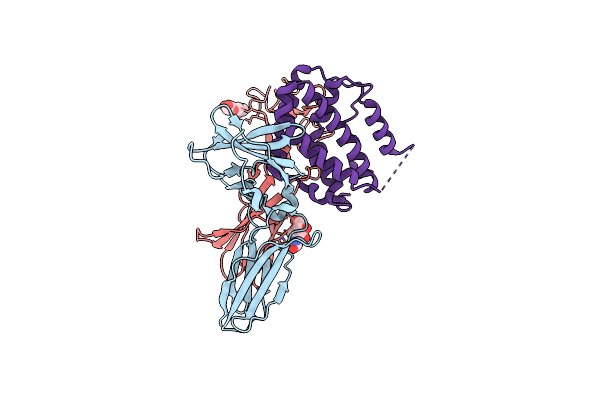 |
Structure Of The Ifn-Lambda3/Ifn-Lambdar1/Il-10Rbeta Receptor Complex With An Engineered Il-10Rbeta
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: CYTOKINE Ligands: NAG |
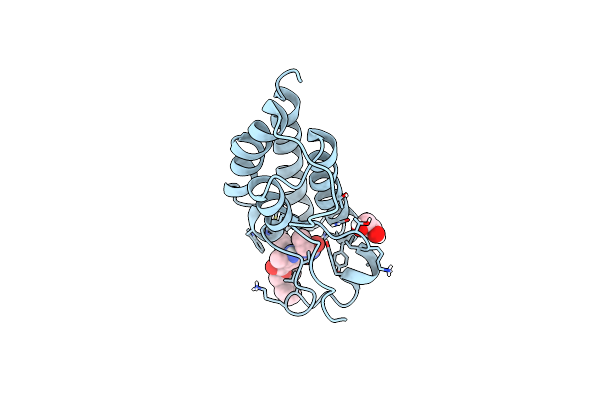 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.22 Å Release Date: 2024-12-18 Classification: TRANSCRIPTION Ligands: YPB, EDO |
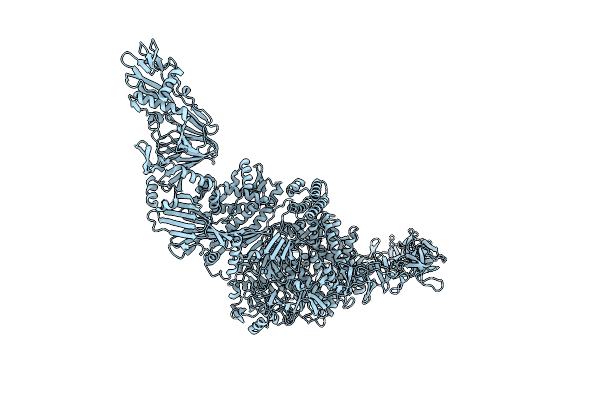 |
Organism: Paeniclostridium sordellii (strain atcc 9714 / dsm 2141 / jcm 3814 / lmg 15708 / ncimb 10717 / 211)
Method: ELECTRON MICROSCOPY Release Date: 2023-12-13 Classification: TOXIN Ligands: ZN |
 |
Organism: Paeniclostridium sordellii (strain atcc 9714 / dsm 2141 / jcm 3814 / lmg 15708 / ncimb 10717 / 211)
Method: ELECTRON MICROSCOPY Release Date: 2023-12-13 Classification: TOXIN Ligands: ZN |
 |
Organism: Paeniclostridium sordellii (strain atcc 9714 / dsm 2141 / jcm 3814 / lmg 15708 / ncimb 10717 / 211)
Method: ELECTRON MICROSCOPY Release Date: 2023-12-13 Classification: TOXIN Ligands: ZN |
 |
Co-Crystal Structure Of A Photosynthetic Lh1-Rc In Complex With Electron Donor Hipip
Organism: Thermochromatium tepidum
Method: X-RAY DIFFRACTION Release Date: 2021-03-03 Classification: PHOTOSYNTHESIS Ligands: HEC, CA, GOL, LHG, PGV, BCL, BPH, UQ8, SO4, FE, MQ8, CRT, CDL, PEF, LMT, SF4 |
 |
Organism: Arthrobacter sp. fb24
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2010-03-09 Classification: OXIDOREDUCTASE Ligands: CU, CA |
 |
Three-Dimensional Structure Of Chymotrypsin Inactivated With (2S) N-Acetyl-L-Alanyl-L-Phenylalanyl-Chloroethyl Ketone: Implications For The Mechanism Of Inactivation Of Serine Proteases By Chloroketones
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1994-11-01 Classification: HYDROLASE(SERINE PROTEINASE) Ligands: HIN |