Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
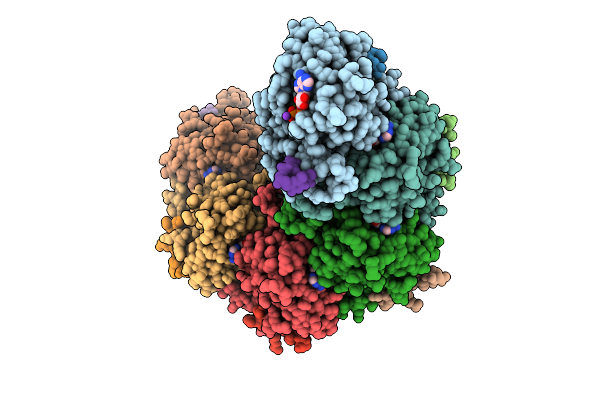 |
Rad51 Filament In Complex With Calcium And Atp Bound By The Rad51Ap1 C-Terminus
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: DNA BINDING PROTEIN Ligands: ATP, CA, K |
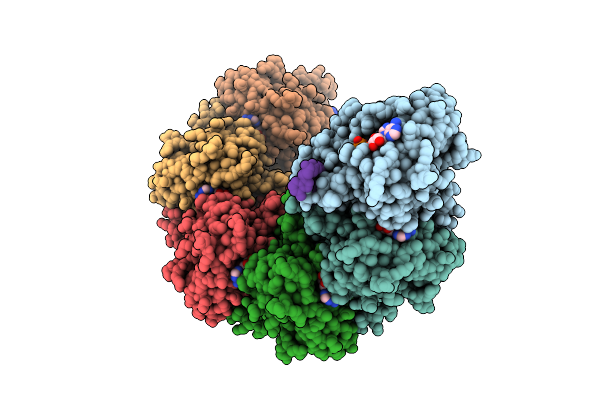 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: DNA BINDING PROTEIN Ligands: ATP, MG, K |
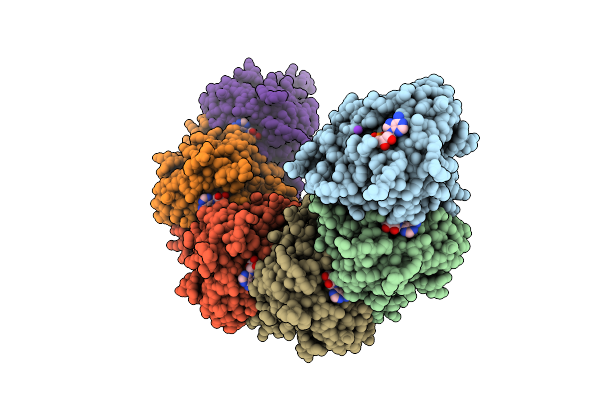 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: DNA BINDING PROTEIN Ligands: MG, ADP, K |
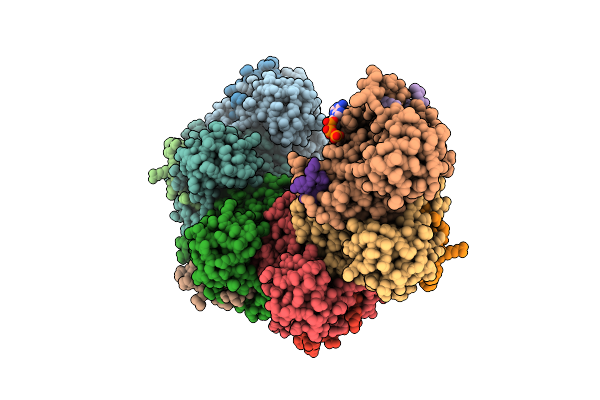 |
Rad51 Filament In Complex With Magnesium And Atp Bound By The Rad51Ap1 C-Terminus
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: DNA BINDING PROTEIN Ligands: ATP, K, MG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-03-27 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-03-20 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-03-20 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-03-20 Classification: MEMBRANE PROTEIN Ligands: A1D5N |
 |
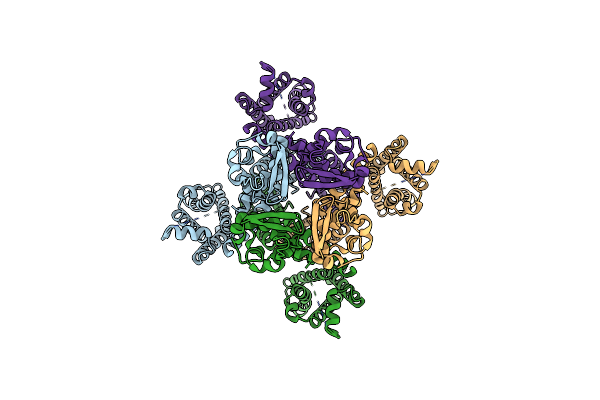
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-03-20 Classification: MEMBRANE PROTEIN |
 |
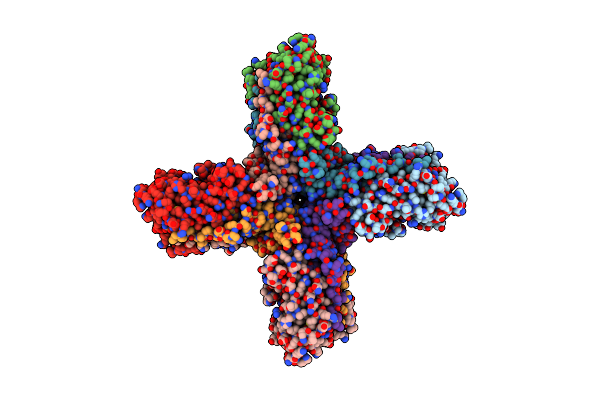
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-11-02 Classification: MEMBRANE PROTEIN |
 |
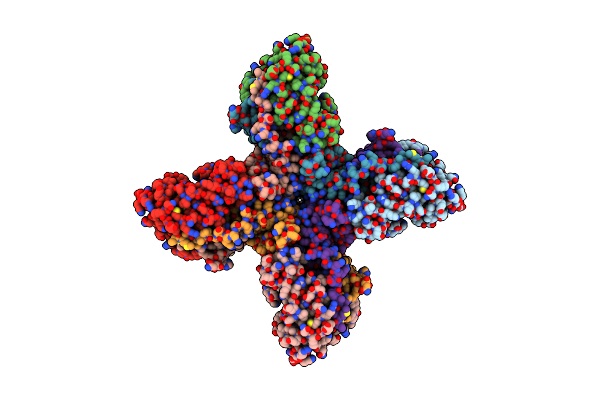
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-11-02 Classification: MEMBRANE PROTEIN |
 |
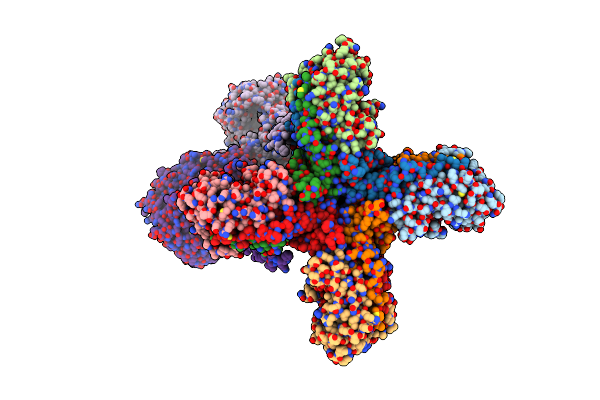
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-11-02 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2022-11-02 Classification: MEMBRANE PROTEIN |
 |
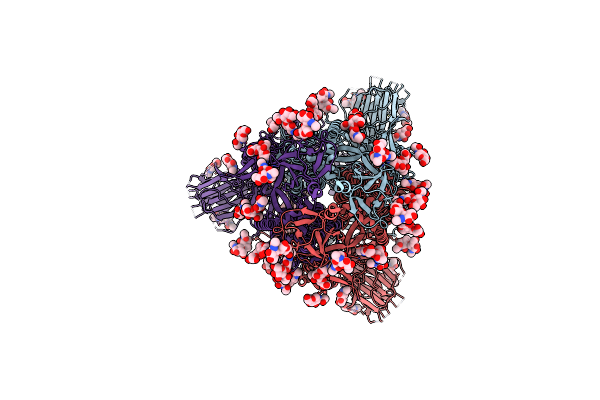
Cryo-Em Structure Of The Sars-Cov-2 Furin Site Mutant S-Trimer From A Subunit Vaccine Candidate
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-03-24 Classification: VIRAL PROTEIN Ligands: NAG, ELA, VCG |
 |
Cryo-Em Structure Of The Sars-Cov-2 Wild-Type S-Trimer From A Subunit Vaccine Candidate
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-03-24 Classification: VIRAL PROTEIN Ligands: NAG, ELA |
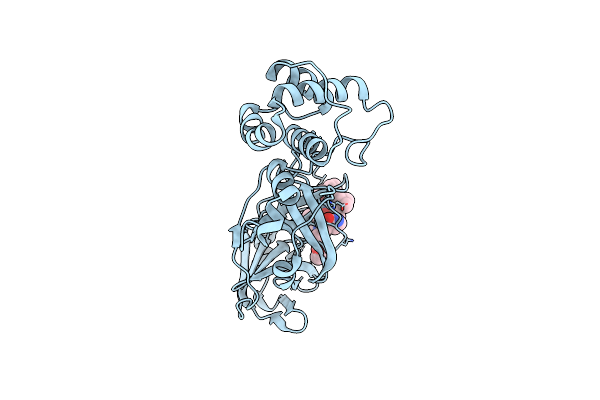 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-06-17 Classification: VIRAL PROTEIN Ligands: TG3 |
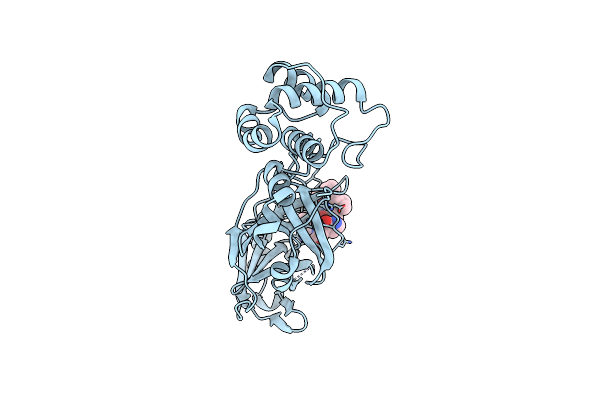 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2020-06-17 Classification: VIRAL PROTEIN Ligands: NOL |
 |
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2019-03-13 Classification: TRANSFERASE Ligands: FAD, MG, EDO |
 |
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2019-03-13 Classification: TRANSFERASE Ligands: FAD, MG |
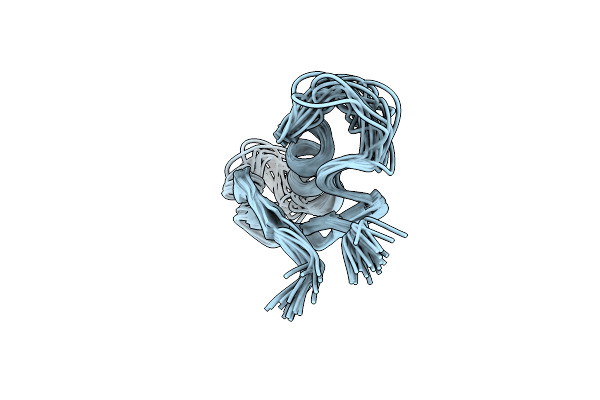 |
Solution Nmr Structure Of Peptide Toxin Sstx From Scolopendra Subspinipes Mutilans
|