Search Count: 18
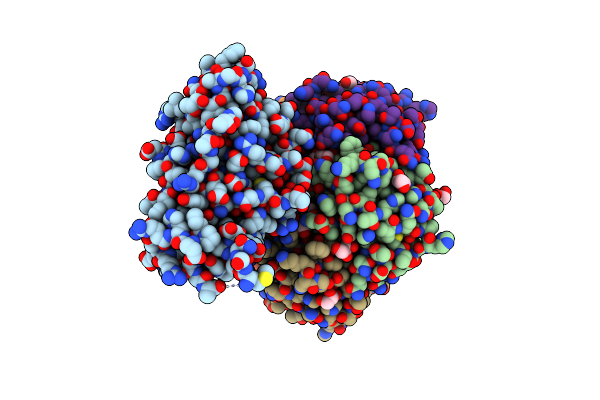 |
Cholera Holotoxin Variant (Chimera With E. Coli Heat-Labile Enterotoxin, 1 C-Terminal Substitution)
Organism: Vibrio cholerae o1
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-02-12 Classification: TOXIN |
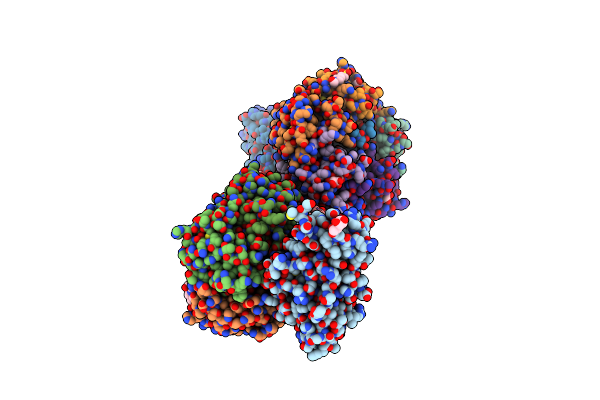 |
Cholera Holotoxin Variant (Chimera With E. Coli Heat-Labile Enterotoxin, 4 C-Terminal Substitutions)
Organism: Vibrio cholerae o1
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-08-14 Classification: TOXIN |
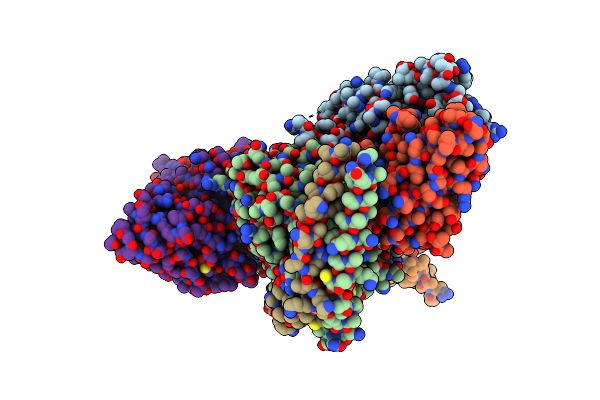 |
Organism: Homo sapiens, Mus musculus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-05-31 Classification: MEMBRANE PROTEIN Ligands: 8IA |
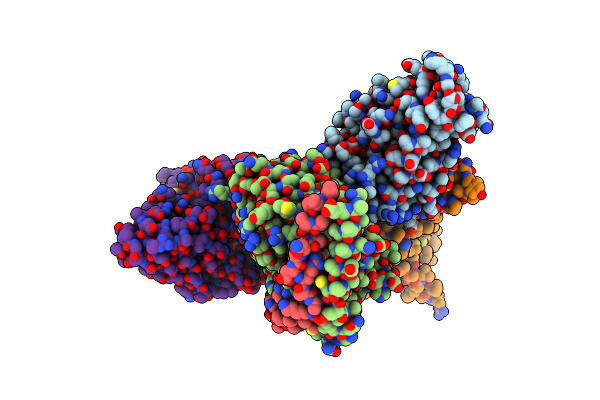 |
Organism: Homo sapiens, Mus musculus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-05-31 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens, Mus musculus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-05-31 Classification: MEMBRANE PROTEIN Ligands: SPD |
 |
Organism: Mus musculus, Homo sapiens, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2023-05-31 Classification: MEMBRANE PROTEIN Ligands: PEA |
 |
Organism: Homo sapiens, Mus musculus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-05-31 Classification: MEMBRANE PROTEIN |
 |
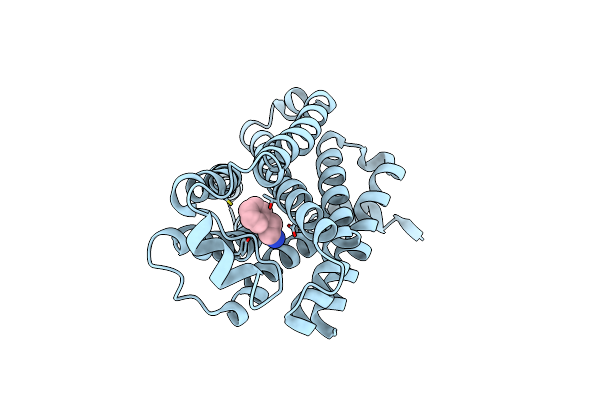
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-05-31 Classification: MEMBRANE PROTEIN Ligands: SPD |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-05-31 Classification: MEMBRANE PROTEIN Ligands: PEA |
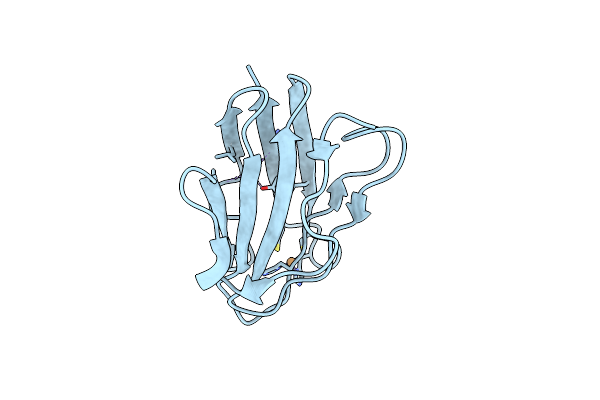 |
Organism: Paracoccus denitrificans
Method: X-RAY DIFFRACTION Resolution:1.09 Å Release Date: 2014-04-23 Classification: ELECTRON TRANSPORT Ligands: CU, NA |
 |
Organism: Paracoccus denitrificans
Method: X-RAY DIFFRACTION Resolution:1.02 Å Release Date: 2014-04-23 Classification: ELECTRON TRANSPORT Ligands: CU1 |
 |
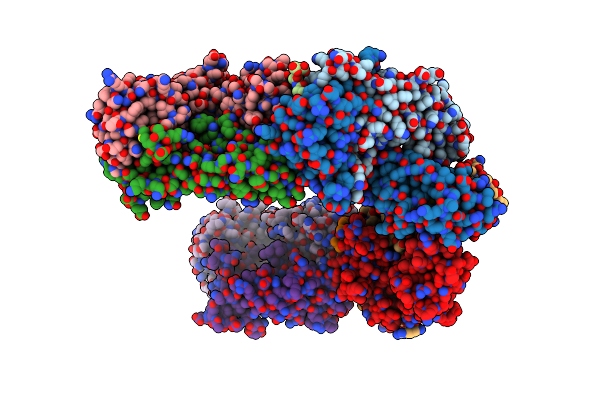
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2013-10-23 Classification: IMMUNE SYSTEM |
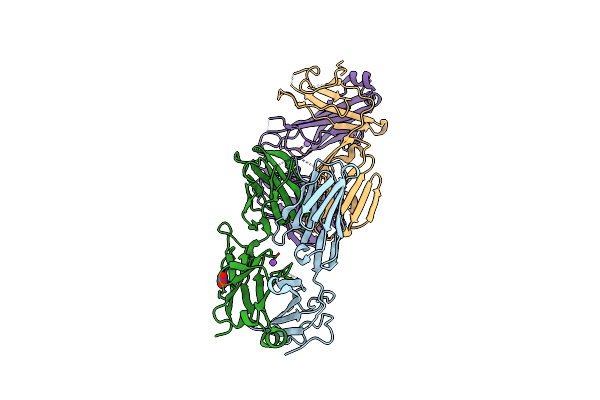
 |
Structural Basis For The Recognition Of Mutant Self By A Tumor-Specific, Mhc Class Ii-Restricted T Cell Receptor G4
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2012-08-29 Classification: IMMUNE SYSTEM |
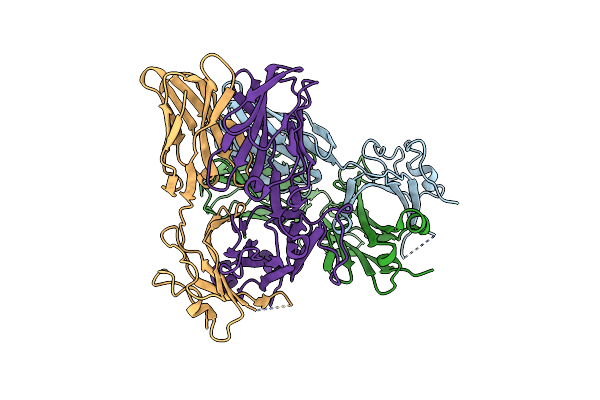
 |
Structural Basis For The Recognition Of Mutant Self By A Tumor-Specific, Mhc Class Ii-Restricted T Cell Receptor G4
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2012-08-29 Classification: IMMUNE SYSTEM Ligands: CL, NO3, NA |
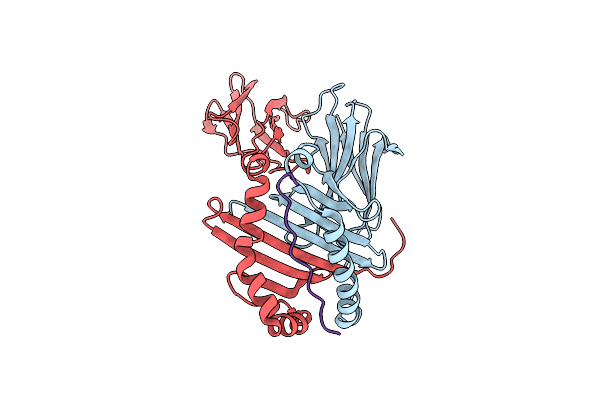 |
Structure Of Mhc Class Ii Molecule Hla-Dr1 Complexed With Phosphopeptide Mart-1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2010-05-12 Classification: IMMUNE SYSTEM/PEPTIDE BINDING PROTEIN |
 |
Structural Basis For Recognition Of Mutant Self By A Tumor-Specific, Mhc Class Ii-Restricted Tcr
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2007-04-03 Classification: IMMUNE SYSTEM |
 |
Structural Basis For Recognition Of Mutant Self By A Tumor-Specific, Mhc Class Ii-Restricted Tcr
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2007-04-03 Classification: IMMUNE SYSTEM |
 |
Structural Basis For Recognition Of Mutant Self By A Tumor-Specific, Mhc Class Ii-Restricted Tcr
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2007-04-03 Classification: IMMUNE SYSTEM |

