Search Count: 16
 |
Organism: Homo sapiens, Tequatrovirus t4
Method: ELECTRON MICROSCOPY Release Date: 2023-05-24 Classification: MEMBRANE PROTEIN Ligands: HXA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-05-24 Classification: MEMBRANE PROTEIN Ligands: 2YB |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-05-24 Classification: MEMBRANE PROTEIN Ligands: 2YB |
 |
Organism: Sepioloidea lineolata
Method: ELECTRON MICROSCOPY Release Date: 2023-04-12 Classification: STRUCTURAL PROTEIN Ligands: WK3 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-06-08 Classification: SIGNALING PROTEIN Ligands: A6F, OLA |
 |
Organism: Homo sapiens, Bos taurus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-05-25 Classification: MEMBRANE PROTEIN Ligands: OLA, A6F |
 |
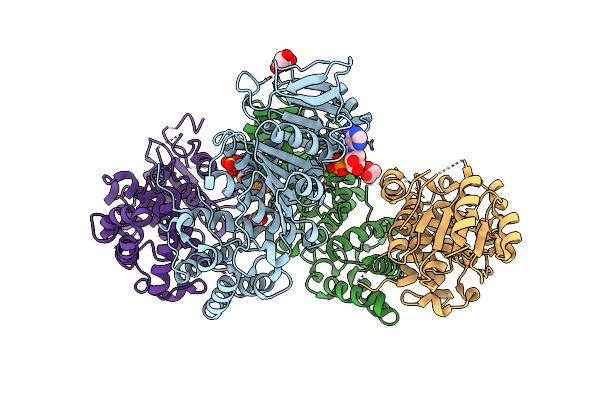
Atomic Model For The N-Terminus Of Trao Fitted In The Full-Length Structure Of The Bacterial Pkm101 Type Iv Secretion System Core Complex
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Resolution:12.40 Å Release Date: 2013-04-03 Classification: MEMBRANE PROTEIN |
 |
Fitting Result In The O-Layer Of The Subnanometer Structure Of The Bacterial Pkm101 Type Iv Secretion System Core Complex Digested With Elastase
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Resolution:8.50 Å Release Date: 2013-04-03 Classification: CELL ADHESION |
 |
Fitting Results In The I-Layer Of The Subnanometer Structure Of The Bacterial Pkm101 Type Iv Secretion System Core Complex Digested With Elastase
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Resolution:8.50 Å Release Date: 2013-04-03 Classification: CELL ADHESION |
 |
Organism: Thermoanaerobacter pseudethanolicus
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2012-07-04 Classification: HYDROLASE Ligands: MG, GOL, ADP, SO4 |
 |
Organism: Thermoanaerobacter pseudethanolicus
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2012-07-04 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:3.14 Å Release Date: 2010-08-11 Classification: HYDROLASE |
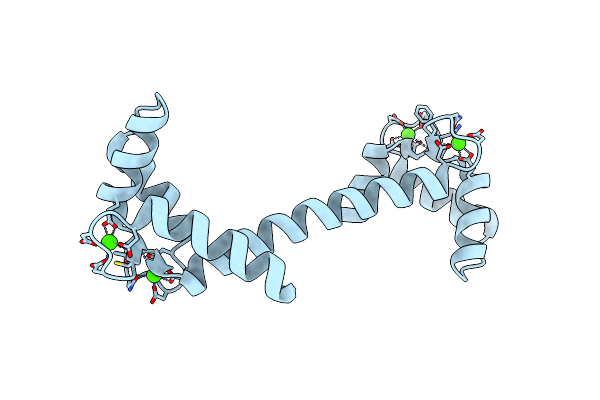 |
Crystal Structure Of The Calmodulin-Like Protein (Hclp) From Human Epithelial Cells
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2002-06-05 Classification: METAL BINDING PROTEIN Ligands: CA |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2002-03-27 Classification: MEMBRANE PROTEIN Ligands: BNG |
 |
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2001-10-31 Classification: ION TRANSPORT Ligands: RET, LI1 |
 |
Bacteriorhodopsin O-Like Intermediate State Of The D85S Mutant At 2.25 Angstrom Resolution
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2001-10-31 Classification: ION TRANSPORT Ligands: RET, LI1 |

