Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
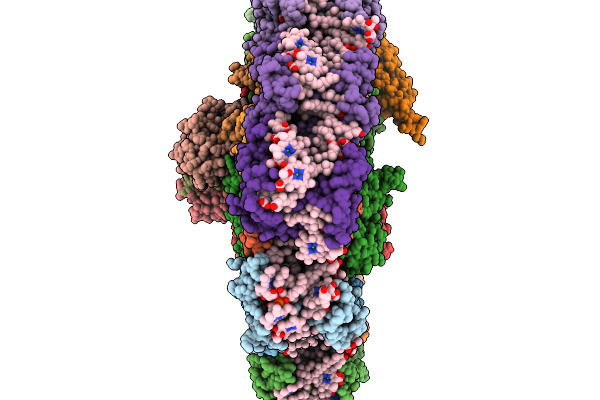 |
Organism: Chroomonas placoidea
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: PHOTOSYNTHESIS Ligands: CLA, KC2, II0, II3, IHT, LHG, LMG, SQD, 8CT, DGD, SF4, PQN |
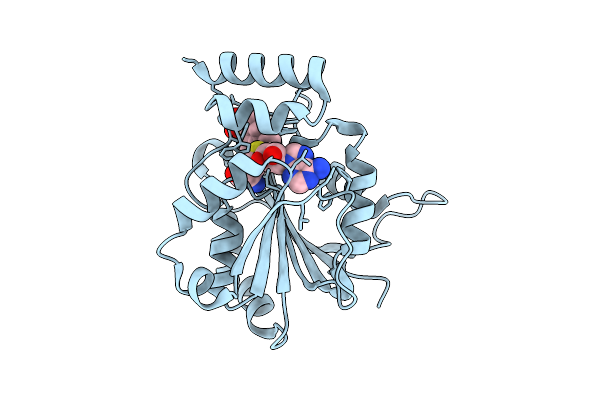 |
Crystal Structure Of Halide Methyltransferase From Tulasnella Calospora In Complex With Sah And Mesitylene Sulfonate
Organism: Tulasnella calospora mut 4182
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: TRANSFERASE Ligands: SAH, A1IRH, CL |
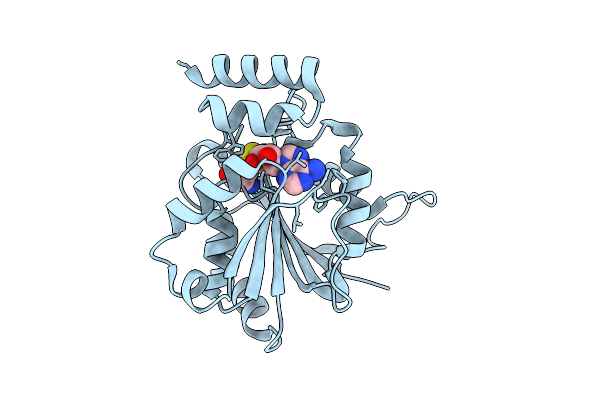 |
Crystal Structure Of Halide Methyltransferase From Tulasnella Calospora In Complex With Sah
Organism: Tulasnella calospora mut 4182
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: TRANSFERASE Ligands: SAH, CL |
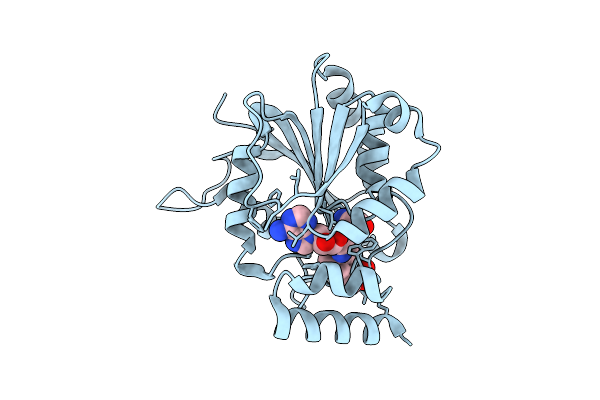 |
Crystal Structure Of Halide Methyltransferase From Tulasnella Calospora In Complex With Sinefungin And Mesitylene Sulfonate
Organism: Tulasnella calospora mut 4182
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: TRANSFERASE Ligands: SFG, A1IRH, CL |
 |
Crystal Structure Of Halide Methyltransferase From Tulasnella Calospora In Complex With Sah And Mesitylene Sulfonate At Room Temperature
Organism: Tulasnella calospora mut 4182
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: TRANSFERASE Ligands: SAH, A1IRH |
 |
Crystal Structure Of Halide Methyltransferase From Tulasnella Calospora In Complex With Sah At Room Temperature
Organism: Tulasnella calospora mut 4182
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: TRANSFERASE Ligands: SAH |
 |
Crystal Structure Of Halide Methyltransferase From Tulasnella Calospora In Complex With Sinefungin And Mesitylene Sulfonate At Room Temperature
Organism: Tulasnella calospora mut 4182
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: TRANSFERASE Ligands: SFG, A1IRH |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: LYASE Ligands: GOL |
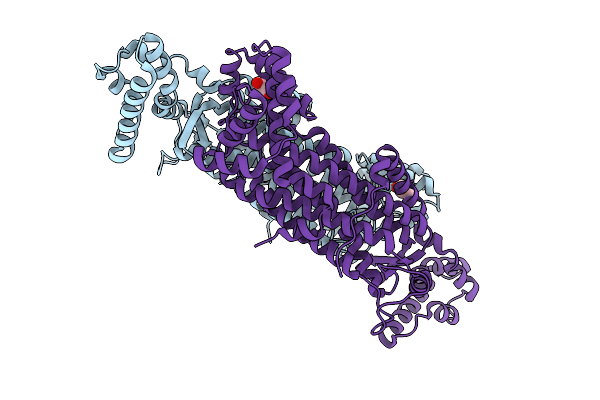 |
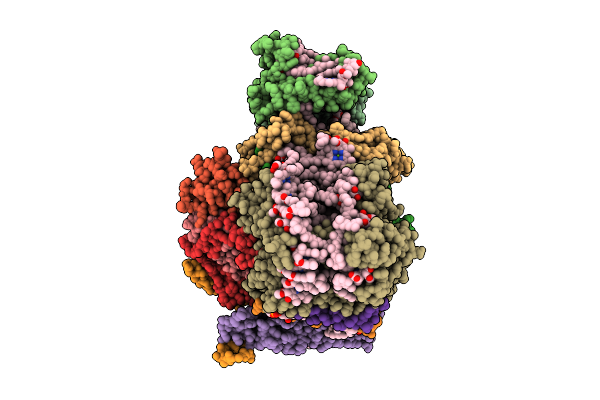
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: LYASE Ligands: GOL |
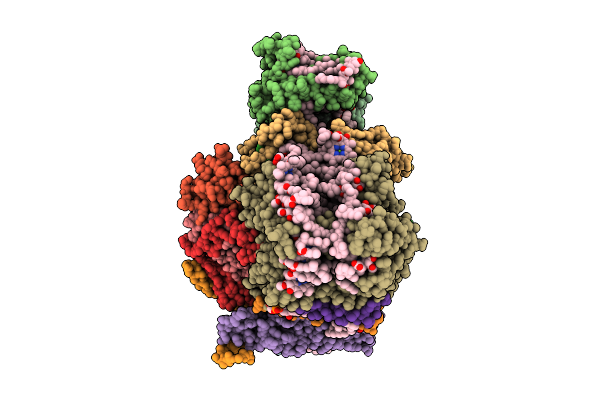 |
Structure Of The Wild-Type Psi-5Vcpi Supercomplex In Nannochloropsis Oceanica
Organism: Nannochloropsis oceanica strain imet1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: XAT, A1L1G, CLA, SQD, DGD, LMG, A1L1F, PQN, LHG, BCR, SF4 |
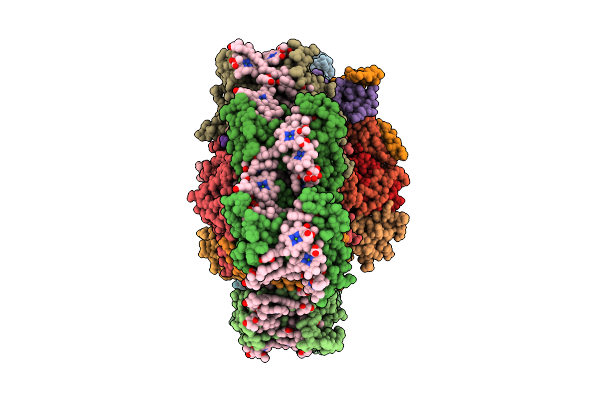 |
Structure Of The Canthaxanthin Mutant Psi-9Vcpi Supercomplex In Nannochloropsis Oceanica
Organism: Nannochloropsis oceanica strain imet1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: XAT, A1L1G, CLA, SQD, A1L1F, 45D, LHG, DGD, LMG, PQN, BCR, SF4 |
 |
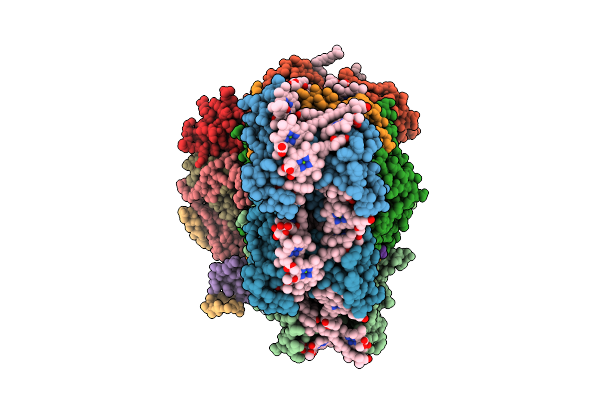
Structure Of The Canthaxanthin Mutant Psi-5Vcpi Supercomplex In Nannochloropsis Oceanica
Organism: Nannochloropsis oceanica strain imet1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: XAT, A1L1G, CLA, SQD, DGD, LMG, A1L1F, PQN, LHG, BCR, SF4 |
 |
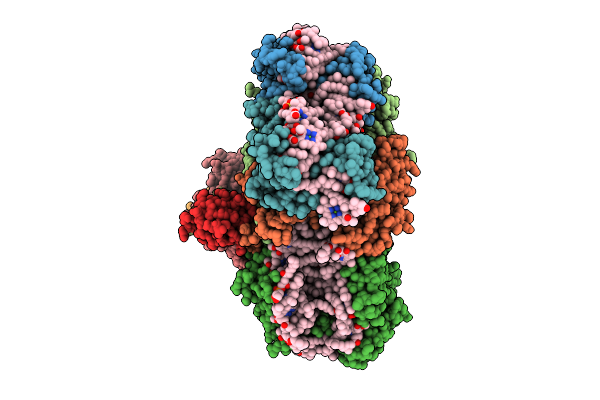
Structure Of The Canthaxanthin Mutant Psi-4Vcpi Supercomplex In Nannochloropsis Oceanica
Organism: Nannochloropsis oceanica strain imet1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: A1L1G, A1L1F, XAT, 45D, LHG, CLA, DGD, SQD, PQN, BCR, SF4, LMG |
 |
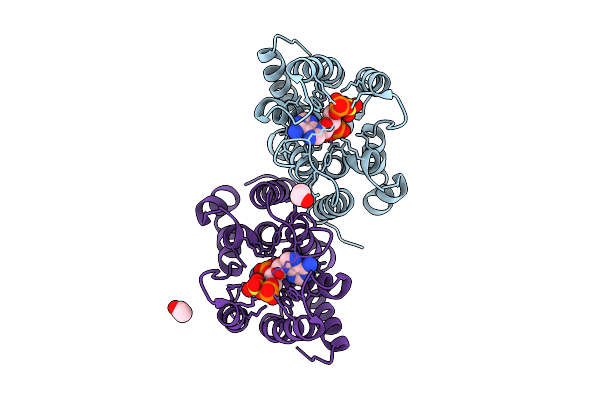
Structure Of The Canthaxanthin Mutant Psi-3Vcpi Supercomplex In Nannochloropsis Oceanica
Organism: Nannochloropsis oceanica strain imet1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: XAT, A1L1G, CLA, SQD, DGD, A1L1F, PQN, LHG, BCR, SF4, LMG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE Ligands: ZN, PPS, EDO, ACY |
 |
Sivtal Integrase In Complex With Rna Stem-Loop (Focused Refinement Of The Filament Repeat Unit)
Organism: Simian immunodeficiency virus, Human immunodeficiency virus 1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN Ligands: ZN |
 |
Cryoem Reconstruction Of Integrase Filament At The Lumen Of Native Hiv-1 Cores (Box Size 34.2 Nm)
Organism: Human immunodeficiency virus 1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN Ligands: ZN, IHP |
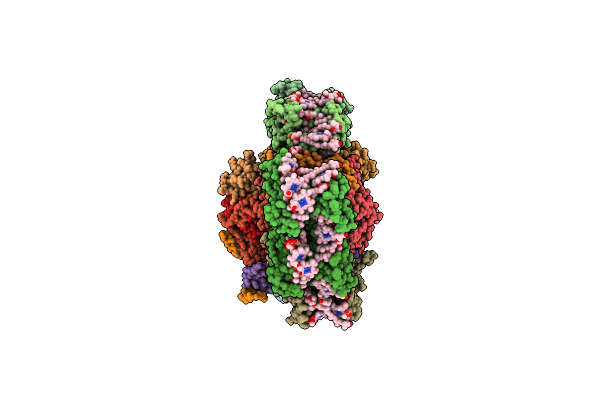 |
Structure Of The Wild-Type Psi-9Vcpi Supercomplex In Nannochloropsis Oceanica
Organism: Nannochloropsis oceanica strain imet1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: PHOTOSYNTHESIS Ligands: XAT, A1L1G, CLA, SQD, A1L1F, LHG, DGD, LMG, PQN, BCR, SF4 |
 |
Structure Of The Wild-Type Psi-8Vcpi Supercomplex In Nannochloropsis Oceanica
Organism: Nannochloropsis oceanica strain imet1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: PHOTOSYNTHESIS Ligands: XAT, A1L1G, CLA, SQD, A1L1F, LHG, DGD, LMG, PQN, BCR, SF4 |