Search Count: 1,632
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: IMMUNE SYSTEM Ligands: CLR, NAG |
 |
Organism: Actinosynnema pretiosum subsp. auranticum
Method: SOLUTION NMR Release Date: 2025-11-26 Classification: ANTIMICROBIAL PROTEIN |
 |
Organism: Escherichia coli k-12, Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN Ligands: A1D6R, NA |
 |
Organism: Escherichia coli k-12, Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN Ligands: A1D6W, NA |
 |
Organism: Acinetobacter baumannii
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: HYDROLASE Ligands: 4BW |
 |
Organism: Acinetobacter baumannii
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: HYDROLASE |
 |
Organism: Acinetobacter baumannii
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: HYDROLASE |
 |
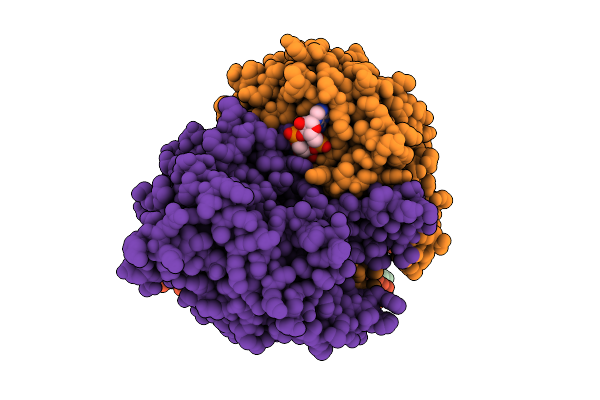
Crystal Structure Of Human Shiftless (Sfl) Containing Phosphorylation Sites Ser249, Thr250, Thr253 And Ser256
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: ANTIVIRAL PROTEIN Ligands: ZN |
 |
Organism: Acinetobacter baumannii
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: HYDROLASE Ligands: 4BW |
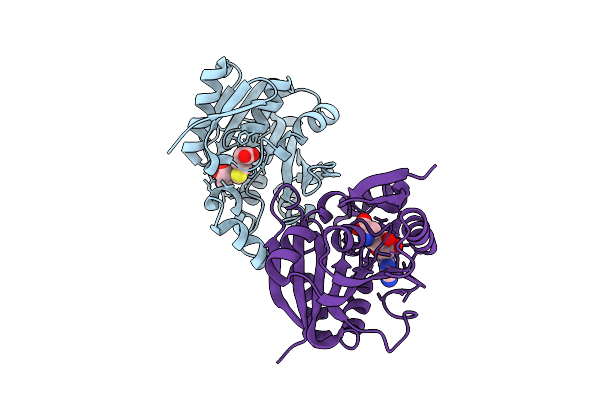 |
Crystal Structure Of The Carbamoyl N-Methyltransferase Asc-Orf2 Complexed With Sah
Organism: Amycolatopsis alba dsm 44262
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: BIOSYNTHETIC PROTEIN Ligands: SAH |
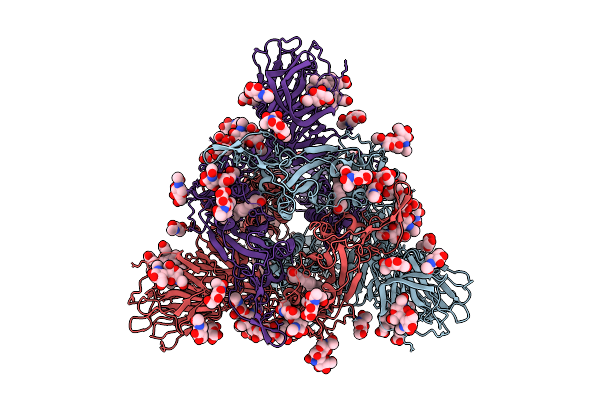 |
Organism: Sarbecovirus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: VIRAL PROTEIN Ligands: BLA, NAG, EIC |
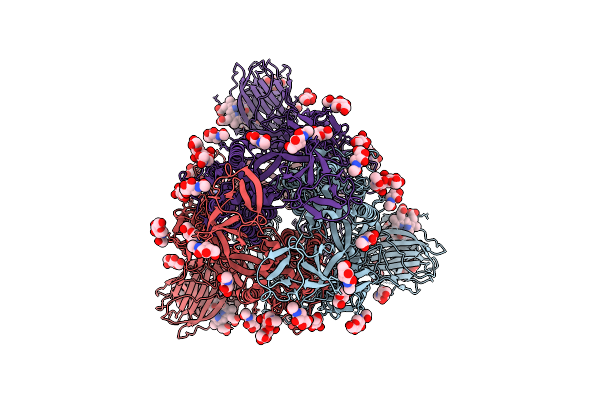 |
Organism: Sarbecovirus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: VIRAL PROTEIN Ligands: NAG, BLA |
 |
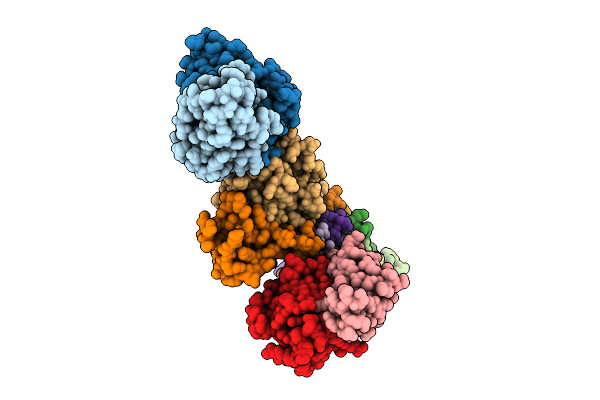
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: IMMUNE SYSTEM Ligands: CLR, LPE |
 |
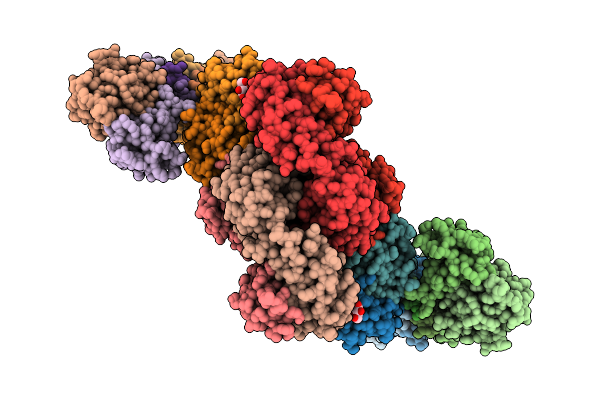
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: IMMUNE SYSTEM Ligands: CLR |
 |
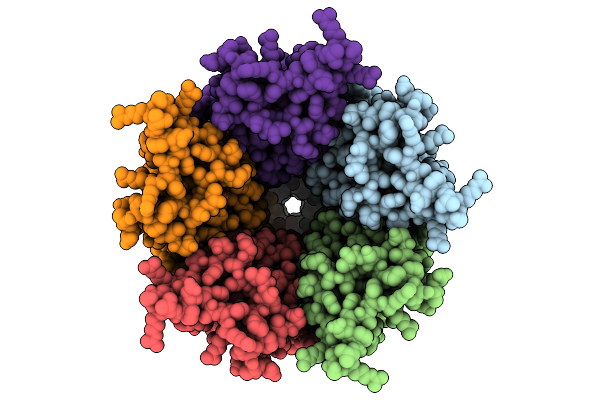
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: IMMUNE SYSTEM Ligands: NAG |
 |
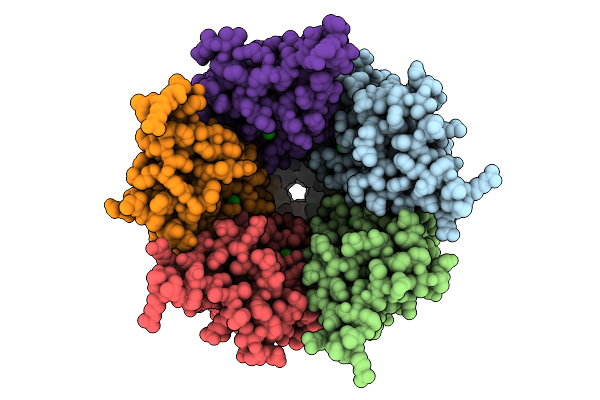
Cryo-Em Structure Of Nanodisc (Pe:Ps:Pc) Reconstituted Glic At Ph 4 In Iiiii Conformation
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: PEE |
 |
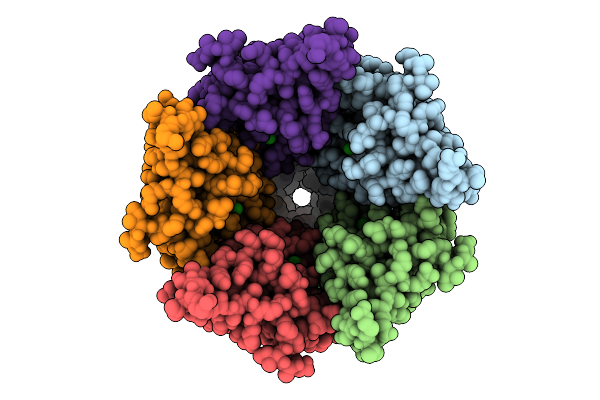
Cryo-Em Structure Of Nanodisc (Pe:Ps:Pc) Reconstituted Glic At Ph 4 In Iiiio Conformation
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: CL, PEE |
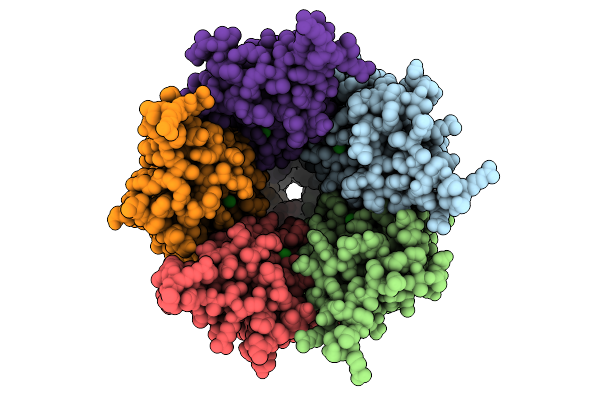 |
Cryo-Em Structure Of Nanodisc (Pe:Ps:Pc) Reconstituted Glic At Ph 4 In Iiioo Conformation
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: CL, PEE |
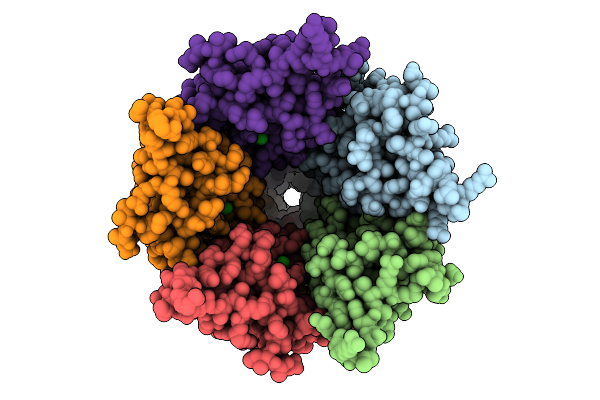 |
Cryo-Em Structure Of Nanodisc (Pe:Ps:Pc) Reconstituted Glic At Ph 4 In Iiooo Conformation
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: CL, PEE |
 |
Cryo-Em Structure Of Nanodisc (Pe:Ps:Pc) Reconstituted Glic At Ph 4 In Ioooo Conformation
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: CL, PEE |

