Search Count: 79
 |
Organism: Anabaena sp. 90
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2024-10-16 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Organism: Anabaena sp. 90
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-10-16 Classification: OXIDOREDUCTASE Ligands: NAD, NAP |
 |
Organism: Anabaena sp. 90
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2024-10-16 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Organism: Anabaena sp. 90
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-10-16 Classification: OXIDOREDUCTASE Ligands: NAD, NAP, FLC |
 |
Organism: Anabaena sp. 90
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-10-16 Classification: OXIDOREDUCTASE Ligands: NAD, FLC |
 |
Organism: Anabaena sp. 90
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-10-16 Classification: OXIDOREDUCTASE Ligands: LMR, GOL, NAD |
 |
Organism: Anabaena sp. 90
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-10-16 Classification: OXIDOREDUCTASE Ligands: LMR, NAD |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2022-05-18 Classification: TRANSFERASE Ligands: SO4 |
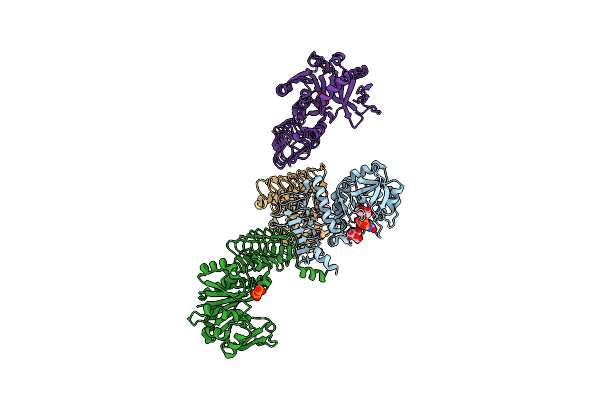 |
Arabidopsis Gdp-D-Mannose Pyrophosphorylase (Vtc1) Structure (Product-Bound)
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2022-05-18 Classification: TRANSFERASE Ligands: GDD, FLC, SO4, PPV |
 |
Structural And Mechanistic Studies Of A Novel Non-Heme Iron Epimerase/Lyase And Its Utilization In Chemoselective Synthesis.
Organism: Streptomyces albus
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2022-03-16 Classification: ISOMERASE Ligands: FE, GOA, 5RN, JAX |
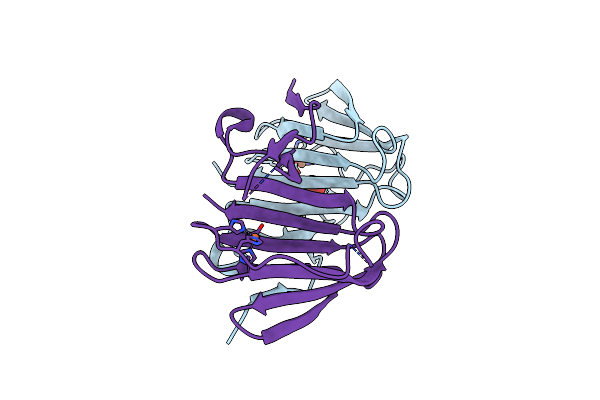 |
Structural And Mechanistic Studies Of A Novel Non-Heme Iron Epimerase/Lyase And Its Utilization In Chemoselective Synthesis.
Organism: Streptomyces albus
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2022-03-16 Classification: ISOMERASE Ligands: PO3, FE |
 |
Structural And Mechanistic Studies Of A Novel Non-Heme Iron Epimerase/Lyase And Its Utilization In Chemoselective Synthesis.
Organism: Streptomyces albus
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2022-03-16 Classification: ISOMERASE Ligands: FE, 3IO |
 |
Structural And Mechanistic Studies Of A Novel Non-Heme Iron Epimerase/Lyase And Its Utilization In Chemoselective Synthesis.
Organism: Streptomyces albus
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2022-03-16 Classification: ISOMERASE Ligands: FE, JCX |
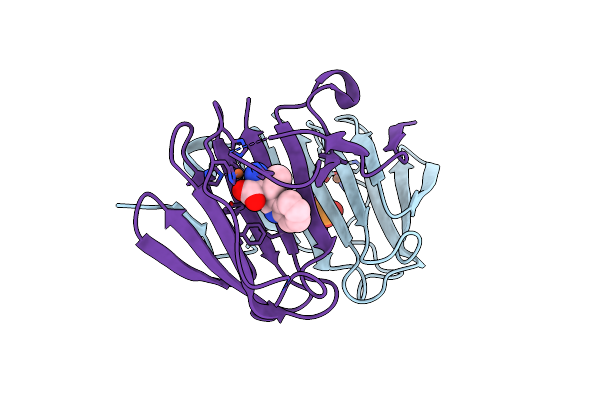 |
Structural And Mechanistic Studies Of A Novel Non-Heme Iron Epimerase/Lyase And Its Utilization In Chemoselective Synthesis.
Organism: Streptomyces albus
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2022-03-16 Classification: ISOMERASE Ligands: PO3, FE, JD3 |
 |
Structural And Mechanistic Studies Of A Novel Non-Heme Iron Epimerase/Lyase And Its Utilization In Chemoselective Synthesis.
Organism: Streptomyces albus
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2022-03-16 Classification: ISOMERASE Ligands: I3A, FE, GOA |
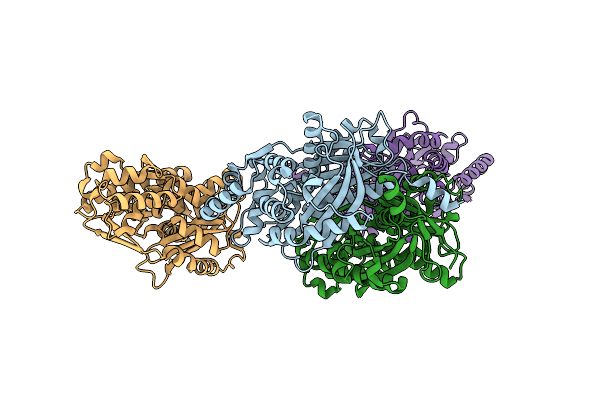 |
Organism: Aloe arborescens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2022-01-19 Classification: TRANSFERASE |
 |
Organism: Rhodopseudomonas palustris
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2021-01-13 Classification: LIGASE |
 |
The Crystal Structure Of Benzoate Coenzyme A Ligase Double Mutant (H333A/I334A) In Complex With 2-Chloro-1,3-Thiazole-5-Carboxylate-Amp
Organism: Rhodopseudomonas palustris
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2021-01-13 Classification: LIGASE Ligands: AMP, F0O |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-10-07 Classification: VIRAL PROTEIN Ligands: GQU, TRS |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2020-09-16 Classification: VIRAL PROTEIN Ligands: GKF, GOL, CL |

