Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: DE NOVO PROTEIN Ligands: EDO, P15 |
 |
Crystal Structure Of Staphylococcus Aureus Ketol-Acid Reductoisomerase In Complex With Mg2+, And Jk-5-114
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Release Date: 2025-04-09 Classification: ISOMERASE Ligands: A1AKJ, MG |
 |
Crystal Structure Of Staphylococcus Aureus Ketol-Acid Reductoisomerase In Complex With Mg2+, And Jk-5-115
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Release Date: 2025-04-09 Classification: ISOMERASE Ligands: MG, A1AKY |
 |
Crystal Structure Of Oryza Sativa Ketol-Acid Reductoisomerase In Complex With Mg2+, And Jk-5-114
Organism: Oryza sativa japonica group
Method: X-RAY DIFFRACTION Release Date: 2025-04-09 Classification: ISOMERASE Ligands: MG, A1AKJ |
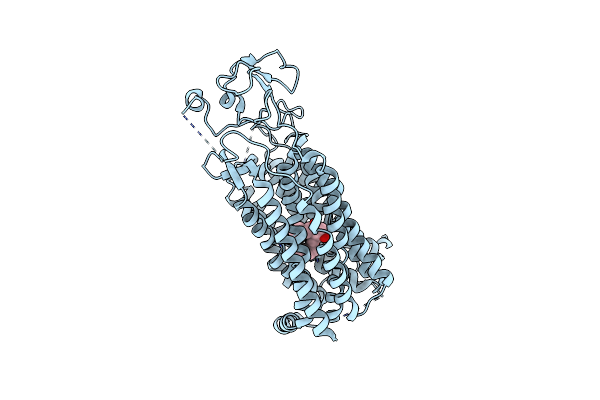 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-09-04 Classification: MEMBRANE PROTEIN Ligands: P2E |
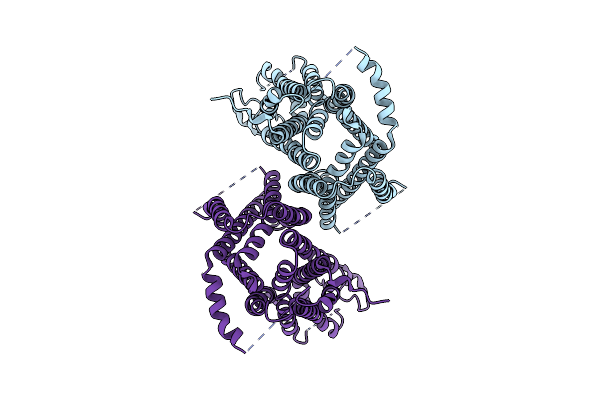 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-21 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-21 Classification: MEMBRANE PROTEIN Ligands: NAG |
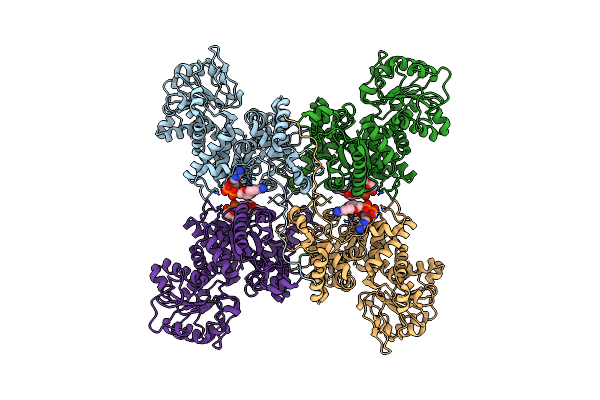 |
Cryo-Em Structures Of Human Mitochondrial Nad(P)+-Dependent Malic Enzyme In Apo Form
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-29 Classification: HYDROLASE Ligands: NAD |
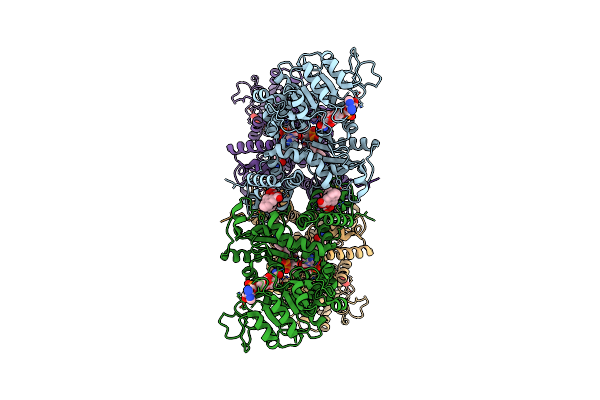 |
Cryo-Em Structures Of Human Mitochondrial Nad(P)+-Dependent Malic Enzyme In A Ternary Complex With Nad+ And Allosteric Inhibitor Ea
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-29 Classification: HYDROLASE Ligands: NAD, JBR |
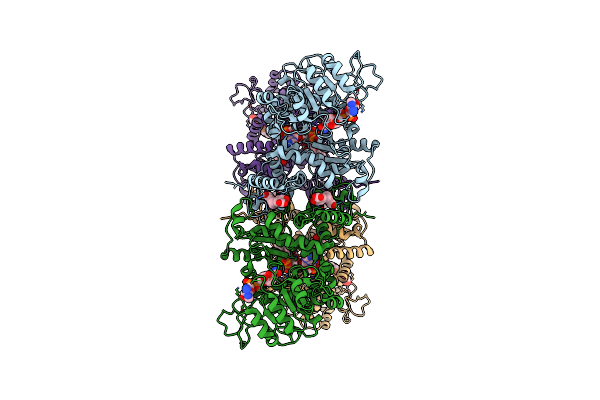 |
Cryo-Em Structures Of Human Mitochondrial Nad(P)+-Dependent Malic Enzyme In A Ternary Complex With Nad+ And Allosteric Inhibitor Mdsa
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-29 Classification: HYDROLASE Ligands: NAD, D5S |
 |
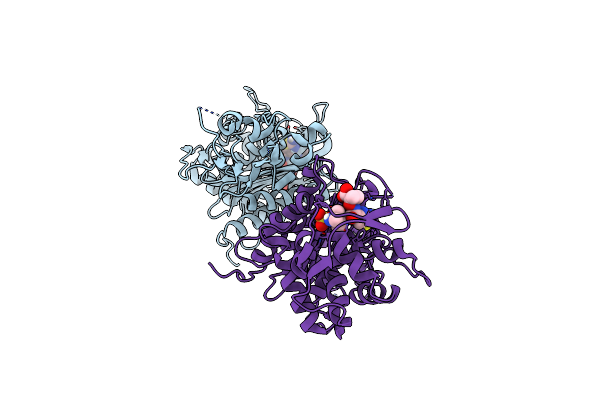
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.97 Å Release Date: 2023-03-29 Classification: DE NOVO PROTEIN |
 |
Structure Of Penicillin-Binding Protein 3 (Pbp3) From Klebsiella Pneumoniae With Ligand 18G
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2022-11-02 Classification: TRANSFERASE Ligands: JXJ |
 |
Organism: Paramecium bursaria chlorella virus 1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-07-06 Classification: BIOSYNTHETIC PROTEIN Ligands: DCM, DCP, ZN, MG |
 |
Chlorovirus Pbcv-1 Bi-Functional Dcmp/Dctp Deaminase Bi-Dcd With Dttp/Dtmp Bound
Organism: Paramecium bursaria chlorella virus 1
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-07-06 Classification: BIOSYNTHETIC PROTEIN Ligands: TTP, TMP, ZN, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2020-10-28 Classification: HYDROLASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2020-10-28 Classification: HYDROLASE Ligands: 95F |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2020-10-28 Classification: HYDROLASE Ligands: 95L |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2020-10-28 Classification: HYDROLASE Ligands: 95R, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-10-28 Classification: HYDROLASE Ligands: C9L |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-10-28 Classification: HYDROLASE Ligands: CEU |