Search Count: 39
 |
Crystal Structure Of Imp-1 Mbl In Complex With 3-(4-Benzyl-1H-1,2,3-Triazol-1-Yl)Phthalic Acid
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2023-06-07 Classification: HYDROLASE/INHIBITOR Ligands: ZN, IT0 |
 |
Crystal Structure Of Imp-1 Mbl In Complex With (3-(4-(P-Tolyl)-1H-1,2,3-Triazol-1-Yl)Benzyl)Phosphonic Acid
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2023-06-07 Classification: HYDROLASE/INHIBITOR Ligands: ZN, ITK |
 |
Crystal Structure Of Vim-2 Mbl In Complex With (2-(4-Phenyl-1H-1,2,3-Triazol-1-Yl)Benzyl)Phosphonic Acid
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2023-06-07 Classification: HYDROLASE/INHIBITOR Ligands: ZN, ITV |
 |
Crystal Structure Of Vim-2 Mbl In Complex With 3-(4-(3-Aminophenyl)-1H-1,2,3-Triazol-1-Yl)Phthalic Acid
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2023-06-07 Classification: HYDROLASE/INHIBITOR Ligands: ZN, IU3 |
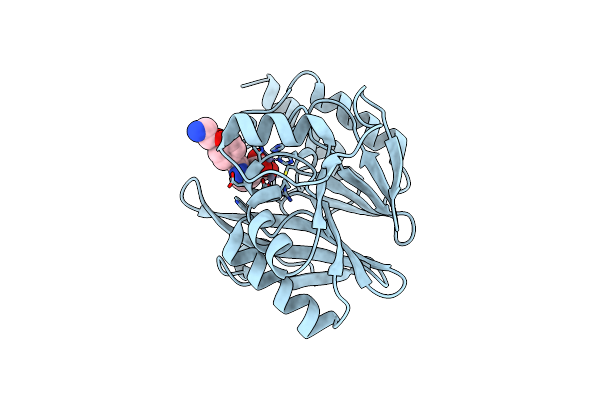 |
Crystal Structure Of Vim-2 Mbl In Complex With 3-(4-(4-(2-Aminoethoxy)Phenyl)-1H-1,2,3-Triazol-1-Yl)Phthalic Acid
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-06-07 Classification: HYDROLASE/INHIBITOR Ligands: ZN, IU7 |
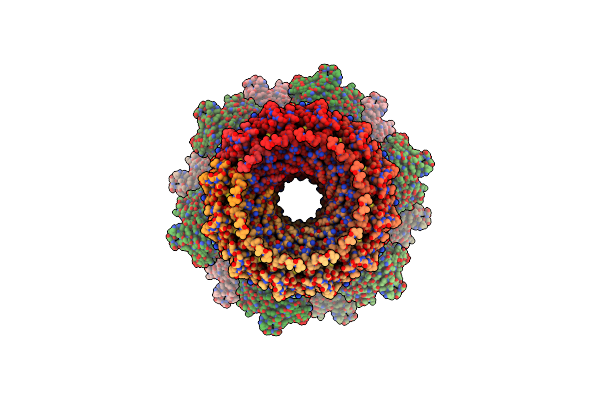 |
Organism: Uncultured cyanophage
Method: ELECTRON MICROSCOPY Release Date: 2023-02-01 Classification: VIRAL PROTEIN |
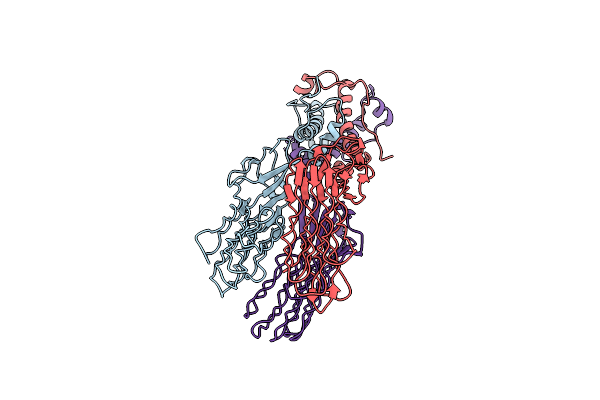 |
Organism: Uncultured cyanophage
Method: ELECTRON MICROSCOPY Release Date: 2023-01-18 Classification: VIRAL PROTEIN |
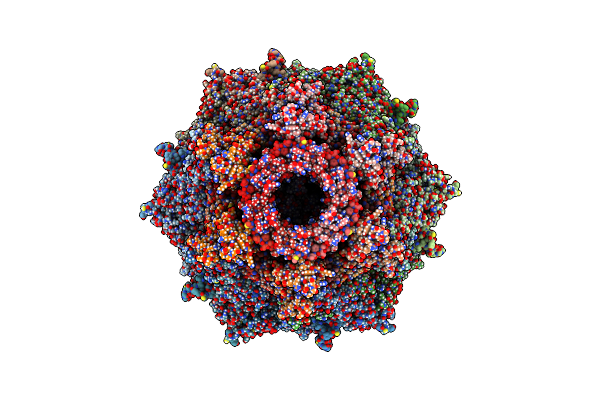 |
Organism: Uncultured cyanophage
Method: ELECTRON MICROSCOPY Release Date: 2023-01-18 Classification: VIRAL PROTEIN |
 |
Organism: Uncultured cyanophage
Method: ELECTRON MICROSCOPY Release Date: 2023-01-18 Classification: VIRAL PROTEIN |
 |
Organism: Uncultured cyanophage
Method: ELECTRON MICROSCOPY Release Date: 2023-01-18 Classification: VIRAL PROTEIN |
 |
Organism: Uncultured cyanophage
Method: ELECTRON MICROSCOPY Release Date: 2023-01-18 Classification: VIRAL PROTEIN |
 |
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2021-10-20 Classification: VIRAL PROTEIN |
 |
 |
 |
Organism: Unidentified
Method: ELECTRON MICROSCOPY Release Date: 2021-10-20 Classification: VIRAL PROTEIN |
 |
Organism: Nostoc sp. (strain pcc 7120 / sag 25.82 / utex 2576)
Method: ELECTRON MICROSCOPY Release Date: 2020-07-15 Classification: LYASE |
 |
Organism: Nostoc sp. (strain pcc 7120 / sag 25.82 / utex 2576)
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2020-05-13 Classification: CHAPERONE/PHOTOSYNTHESIS |
 |
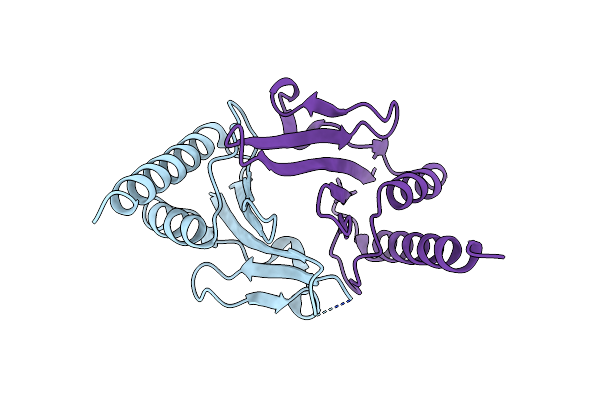
Crystal Structure Of Rubisco Accumulation Factor Raf1 From Anabaena Sp. Pcc 7120
Organism: Nostoc sp. (strain pcc 7120 / sag 25.82 / utex 2576)
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2020-05-13 Classification: CHAPERONE |
 |
Crystal Structure Of A Novel Fold Protein Gp72 From The Freshwater Cyanophage Mic1
Organism: Microcystis phage mic1
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2020-05-13 Classification: STRUCTURAL PROTEIN |
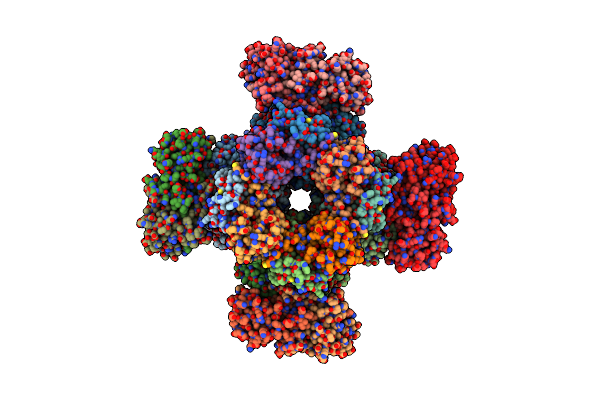 |
Organism: Nostoc sp. (strain pcc 7120 / sag 25.82 / utex 2576)
Method: ELECTRON MICROSCOPY Release Date: 2020-05-13 Classification: CHAPERONE/LYASE |

