Search Count: 136
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.70 Å Release Date: 2026-01-14 Classification: TRANSCRIPTION Ligands: A1CNQ |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.20 Å Release Date: 2026-01-14 Classification: LIGASE Ligands: ZN, A1CN1 |
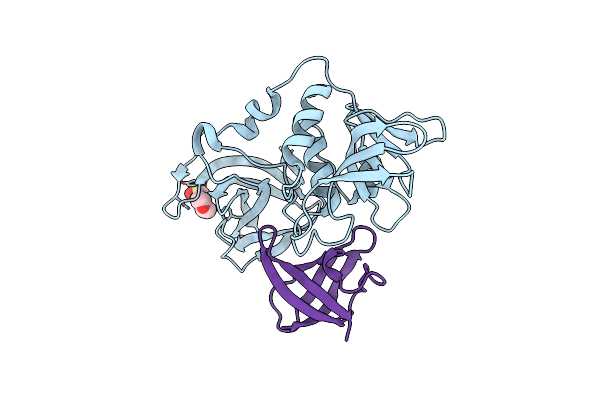 |
Crystal Structure Of Holo-Plygrcs, A Bacteriophage Endolysin In Complex With Cold Shock Protein C
Organism: Staphylococcus phage grcs, Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2025-01-01 Classification: ANTIMICROBIAL PROTEIN Ligands: MES |
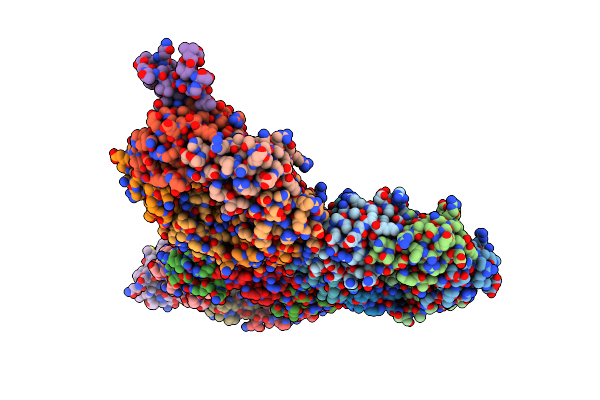 |
Cryo-Em Structure Of Carboxysomal Midi-Shell:Icosahedral Assembly From Csos4A/4B/1A/1B/1C/1D And Csos2 C-Terminal Co-Expression (T = 19)
Organism: Halothiobacillus neapolitanus
Method: ELECTRON MICROSCOPY Release Date: 2024-12-11 Classification: STRUCTURAL PROTEIN |
 |
Cryo-Em Structure Of Carboxysomal Midi-Shell: Icosahedral Assembly From Csos4A/4B/1A/1B/1C/1D And Csos2 C-Terminal Co-Expression (T = 16)
Organism: Halothiobacillus neapolitanus
Method: ELECTRON MICROSCOPY Release Date: 2024-12-11 Classification: STRUCTURAL PROTEIN |
 |
Cryo-Em Structure Of Carboxysomal Midi-Shell: Icosahedral Assembly From Csos4A/4B/1A/1B/1C/1D And Csos2 C-Terminal Co-Expression (T = 9)
Organism: Halothiobacillus neapolitanus
Method: ELECTRON MICROSCOPY Release Date: 2024-12-11 Classification: STRUCTURAL PROTEIN |
 |
Cryo-Em Structure Of Carboxysomal Midi-Shell: Assembly From Csos4A/4B/1A/1B/1C/1D And Csos2 C-Terminal Co-Expression (T=9 Q=12)
Organism: Halothiobacillus neapolitanus
Method: ELECTRON MICROSCOPY Release Date: 2024-12-11 Classification: STRUCTURAL PROTEIN |
 |
Cryo-Em Structure Of Carboxysomal Midi-Shell: Icosahedral Assembly From Csos4A/4B/1A/1B/1C/1D And Csos2 C-Terminal Co-Expression (T = 13)
Organism: Halothiobacillus neapolitanus
Method: ELECTRON MICROSCOPY Release Date: 2024-12-11 Classification: STRUCTURAL PROTEIN |
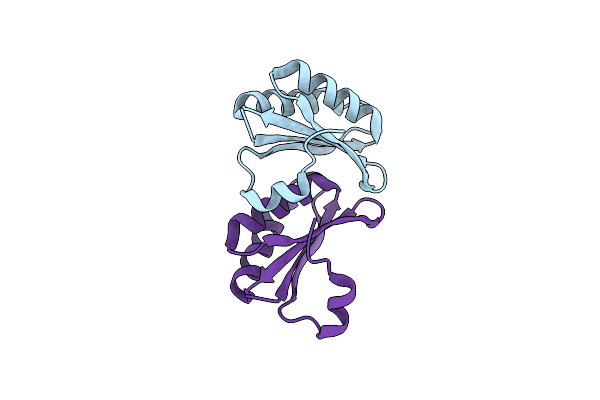 |
Organism: Halothiobacillus neapolitanus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-12-11 Classification: STRUCTURAL PROTEIN |
 |
Cryo-Em Structure Of Carboxysomal Mini-Shell Icosahedral Assembly From Co-Expression Of Csos1C, Csos4A, And Csos2-C (T = 9)
Organism: Halothiobacillus neapolitanus
Method: ELECTRON MICROSCOPY Release Date: 2024-12-11 Classification: STRUCTURAL PROTEIN Ligands: MG |
 |
Structure Of Amplified Asyn Filament By Using Seed Amplification Assay (Saa) From Msa Patient Csf.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-23 Classification: PROTEIN FIBRIL |
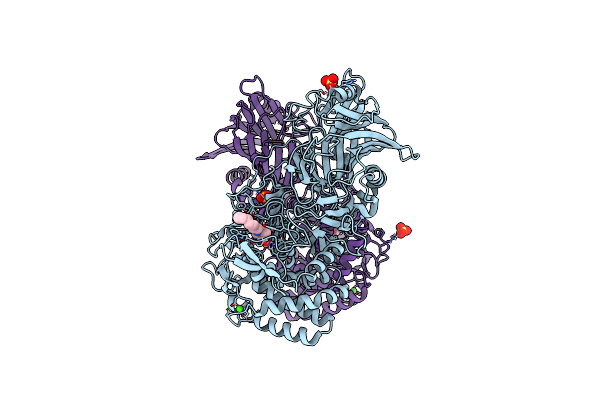 |
Organism: Thermoanaerobacterium xylanolyticum lx-11
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-10-02 Classification: HYDROLASE Ligands: CA, CL, SO4, XHR |
 |
High Resolution Crystal Structure Of A Leaf-Branch Compost Cutinase Quadruple Variant
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2024-09-04 Classification: HYDROLASE Ligands: DIO |
 |
Structure Of P110 Alpha Bound To (S)-1-(4-((2-(4-(4-(2-Amino-4-(Difluoromethyl)Pyrimidin-5-Yl)-6-(3-Methylmorpholino)-1,3,5- Triazin-2-Yl)Piperazin-1-Yl)-2-Oxoethoxy)Methyl)Piperidin-1-Yl)Prop-2-En-1-One (Compound 9)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2024-08-28 Classification: TRANSFERASE/Inhibitor Ligands: RNX |
 |
Organism: Lachnospira eligens
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2024-07-24 Classification: HYDROLASE Ligands: WOF, WR8, GOL |
 |
Structural Basis For The Distinct Roles Of Non-Conserved Pro116 And Conserved Tyr124 Of Bch Domain Of Yeast P50Rhogap
Organism: Schizosaccharomyces pombe 972h-
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2024-05-29 Classification: LIPID BINDING PROTEIN Ligands: PG4 |
 |
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:4.70 Å Release Date: 2024-04-24 Classification: SIGNALING PROTEIN Ligands: NAG |
 |
Organism: Lachnospira eligens
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2024-03-20 Classification: HYDROLASE Ligands: ZBL, GOL |
 |
Organism: Lachnospira eligens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-03-20 Classification: HYDROLASE Ligands: ZG5 |
 |
Organism: Roseburia hominis
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2024-03-20 Classification: HYDROLASE Ligands: GOL, BDP, FMN, ZG5 |

