Search Count: 149
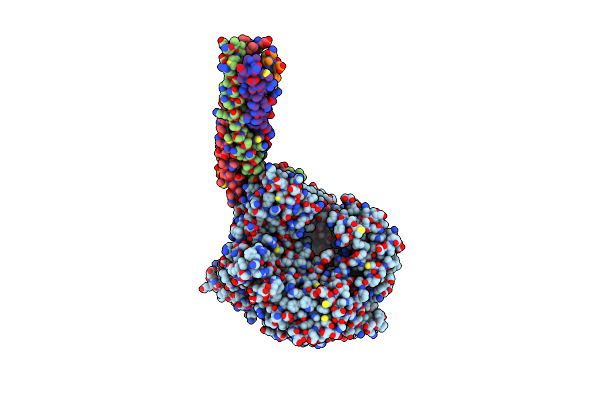 |
Structure Of The Mumps Virus L Protein (State2) Bound By Phosphoprotein Tetramer
Organism: Mumps virus strain jeryl lynn
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: VIRAL PROTEIN Ligands: ZN |
 |
Organism: Streptomyces luteocolor
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2023-05-31 Classification: OXIDOREDUCTASE Ligands: FAD, GLY |
 |
Organism: Streptomyces luteocolor
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-05-31 Classification: OXIDOREDUCTASE Ligands: FAD, I55, GLY |
 |
Organism: Streptomyces luteocolor
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2023-05-31 Classification: OXIDOREDUCTASE Ligands: FAD, I55, GLY |
 |
Organism: Streptomyces luteocolor
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2023-05-31 Classification: OXIDOREDUCTASE Ligands: FAD, SAR |
 |
Organism: Streptomyces luteocolor
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2023-05-31 Classification: OXIDOREDUCTASE Ligands: FAD, GLY, I60 |
 |
Organism: Streptomyces luteocolor
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2023-05-31 Classification: OXIDOREDUCTASE Ligands: FAD, GLY, OI9 |
 |
Organism: Streptomyces luteocolor
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-05-31 Classification: OXIDOREDUCTASE Ligands: FAD, I55 |
 |
Organism: Streptomyces luteocolor
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2023-05-31 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Organism: Streptomyces luteocolor
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-05-31 Classification: OXIDOREDUCTASE Ligands: FAD, GLY, I55 |
 |
Organism: Streptomyces luteocolor
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2023-05-31 Classification: OXIDOREDUCTASE Ligands: FAD, I55, GLY |
 |
Organism: Streptomyces lavendulae subsp. lavendulae
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2023-05-31 Classification: TRANSFERASE Ligands: I55, COA |
 |
Organism: Streptomyces lavendulae subsp. lavendulae
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2023-05-31 Classification: TRANSFERASE Ligands: OI9 |
 |
Organism: Streptomyces luteocolor
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2023-05-31 Classification: OXIDOREDUCTASE Ligands: FAD, GLY, I55 |
 |
Organism: Streptomyces kasugaensis
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2022-11-16 Classification: ISOMERASE |
 |
The Crystal Structure Of Non-Hydrolyzing Udpglcnac 2-Epimerase In Complex With Udp-Glucose
Organism: Streptomyces kasugaensis
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2022-11-16 Classification: ISOMERASE Ligands: UPG, NA |
 |
The Crystal Structure Of Non-Hydrolyzing Udpglcnac 2-Epimerase In Complex With Udp
Organism: Streptomyces kasugaensis
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2022-11-16 Classification: ISOMERASE Ligands: UDP, NA |
 |
Organism: Saccharothrix mutabilis subsp. capreolus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-05-11 Classification: TRANSFERASE |
 |
Organism: Saccharothrix mutabilis subsp. capreolus
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2022-05-11 Classification: TRANSFERASE Ligands: ATP |
 |
Crystal Structure Of Capreomycin Phosphotransferase In Complex With Cmn Iia
Organism: Saccharothrix mutabilis subsp. capreolus
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2022-05-11 Classification: TRANSFERASE |

