Search Count: 2,890
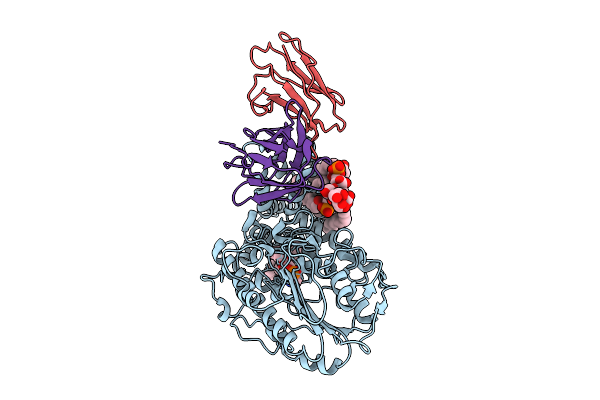 |
Single-Particle Cryo-Em Structure Of The First Variant Of Mobilized Colistin Resistance (Mcr-1) In Its Ligand-Bound State
Organism: Escherichia coli, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSFERASE Ligands: KDL, PEE |
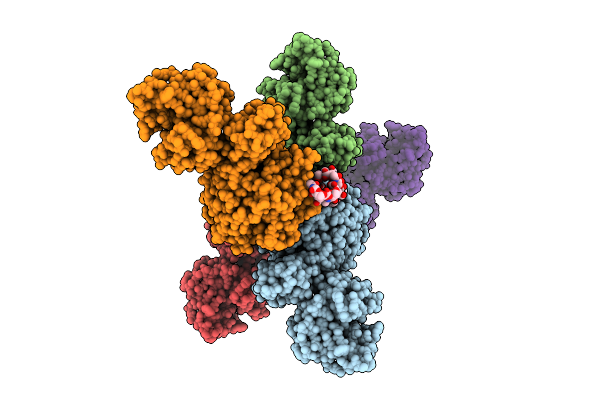 |
Organism: Streptomyces coelicolor
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TOXIN Ligands: A2G, A1CAY, A1CAZ |
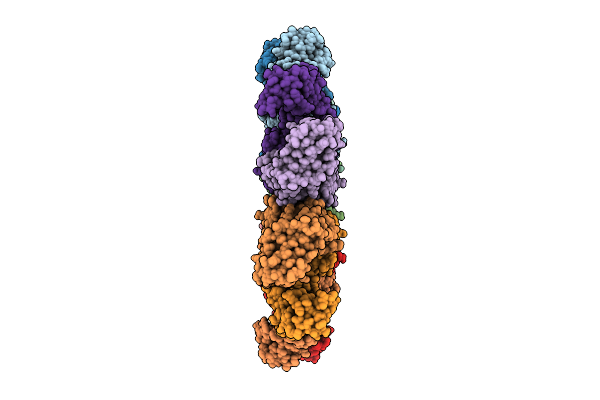 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: IMMUNE SYSTEM |
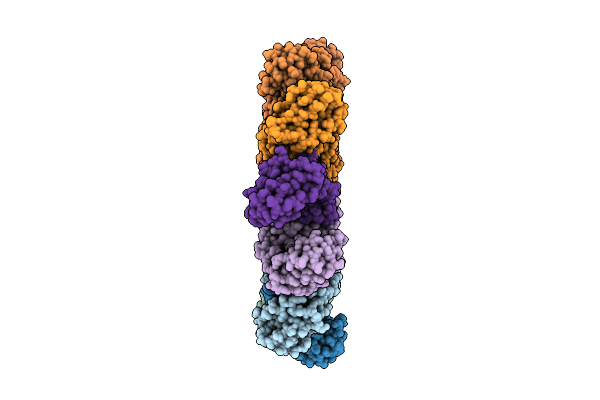 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: IMMUNE SYSTEM |
 |
Organism: Xenopus laevis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: IMMUNE SYSTEM |
 |
Organism: Archangium gephyra
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: CELL INVASION |
 |
Organism: Archangium gephyra
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: CELL INVASION Ligands: Y43, ZN, NCA |
 |
Organism: Archangium gephyra
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: CELL INVASION |
 |
Organism: Archangium gephyra
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: CELL INVASION |
 |
Organism: Archangium gephyra
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: CELL INVASION |
 |
Organism: Archangium gephyra
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: CELL INVASION Ligands: Y43, ZN |
 |
Cryo-Em Structure Of Sars-Cov-2 Ba.5 Spike Protein In Complex With Nab 1C4 (Local Refinement)
Organism: Severe acute respiratory syndrome coronavirus 2, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Cryo-Em Structure Of Sars-Cov-2 Spike Protein In Complex With Three-Nab 8H12, 3E2 And 1C4
Organism: Severe acute respiratory syndrome coronavirus 2, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Cryo-Em Structure Of Sars-Cov-2 Ba.1 Spike Protein In Complex With Three-Nab 8H12, 3E2 And 1C4
Organism: Severe acute respiratory syndrome coronavirus 2, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN |
 |
Cryo-Em Structure Of Sars-Cov-2 Ba.2 Spike Protein In Complex With Triple-Nab 8H12, 3E2 And 1C4 (Local Refinement)
Organism: Severe acute respiratory syndrome coronavirus 2, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Cryo-Em Structure Of Hiv-1 Bg505Ds-Sosip.664 Env Trimer Bound To Dfph-A.01_10R59P_Lc Fab
Organism: Human immunodeficiency virus 1, Macaca
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Streptomyces monomycini
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: HEM |
 |
Organism: Streptomyces monomycini
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: HEM, ARG |

